Pancreatic cancer drug tolerance detection method and kit
A detection kit and detection method technology, applied in the field of genetic engineering, can solve problems such as drug resistance, and achieve the effects of high specificity, low cost, and high prediction accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] Pancreatic cancer clinical drug Gemze drug-resistant cell line obtained
[0026] Pancreatic cancer cells PANC-1 and MIA PaCa-2 were screened for drug-resistant cell lines using gemcitabin (Gemzar), a standard chemotherapy drug for pancreatic cancer. Through drug dose-escalation screening method, PANC-1 and MIA PaCa-2 drug-resistant cell lines that can survive in 56.4 μM and 15.0 μM Gemzar respectively were obtained. The semi-lethal inhibitory concentration (IC50) analysis showed that the IC50 of the drug-resistant cell lines PANC-1-R and MIA PaCa-2-R was increased by 121 times and 6 times, respectively, compared with the drug-sensitive cell lines. The increase in the expression of related drug resistance proteins is a key factor for cancer to acquire drug resistance. For example, MRP1 and MDR1 are upregulated to varying degrees in the acquisition of drug resistance of most cancers. Therefore, we further detected the changes of PANC-1-R (PR) and MIA PaCa-2-R (MR) resist...
Embodiment 2
[0035] RNA Extraction from Pancreatic Cancer and Gemzeitan Drug-resistant Tissues
[0036] 1) Grind the tissue with liquid nitrogen, add 1mL Trizo to each sample, lyse, let it stand at room temperature for 5min, and transfer it to a 1.5mL centrifuge tube;
[0037] 2) Add 200 μL of chloroform, shake fully or turn it upside down several times, after the lysate turns pink, let stand at room temperature for 5 minutes;
[0038] 3) Centrifuge at 12,000g for 15min at 4°C, absorb the supernatant and put it into another 1.5ml centrifuge tube; add 500μL isopropanol, mix the liquid in the tube upside down, and let stand at room temperature for 10min;
[0039] 4) Centrifuge at 12,000g for 10min at 4°C, discard the supernatant;
[0040] 5) Add 1mL of 75% ethanol to slowly wash the white precipitate; centrifuge at 8,000g for 10min at 4°C, discard the supernatant;
[0041] 6) Dry in the air, add an appropriate amount of ultrapure water (RNase-free) to dissolve the precipitate, which can be...
Embodiment 3
[0043] Target miRNA detection in pancreatic cancer and Gemzeita drug-resistant samples
[0044] To detect miR-584, miR-153, miR-188, and miR-483 in the RNA extracted in Example 2, the primers used are as follows:
[0045]
[0046] 1. Reverse transcription PCR:
[0047] Reverse transcription PCR (RT-PCR)
[0048]
[0049] Reverse transcription program: Specific primers: 42°C for 15min, 85°C for 5sec, 4°C to terminate the reaction.
[0050] The cDNA used in the next step for fluorescent quantitative PCR amplification was obtained by reverse transcription PCR.
[0051] 2. Real-time fluorescent quantitative PCR:
[0052] Real-time fluorescence quantitative PCR (qRT-PCR) reaction system (20μL)
[0053]
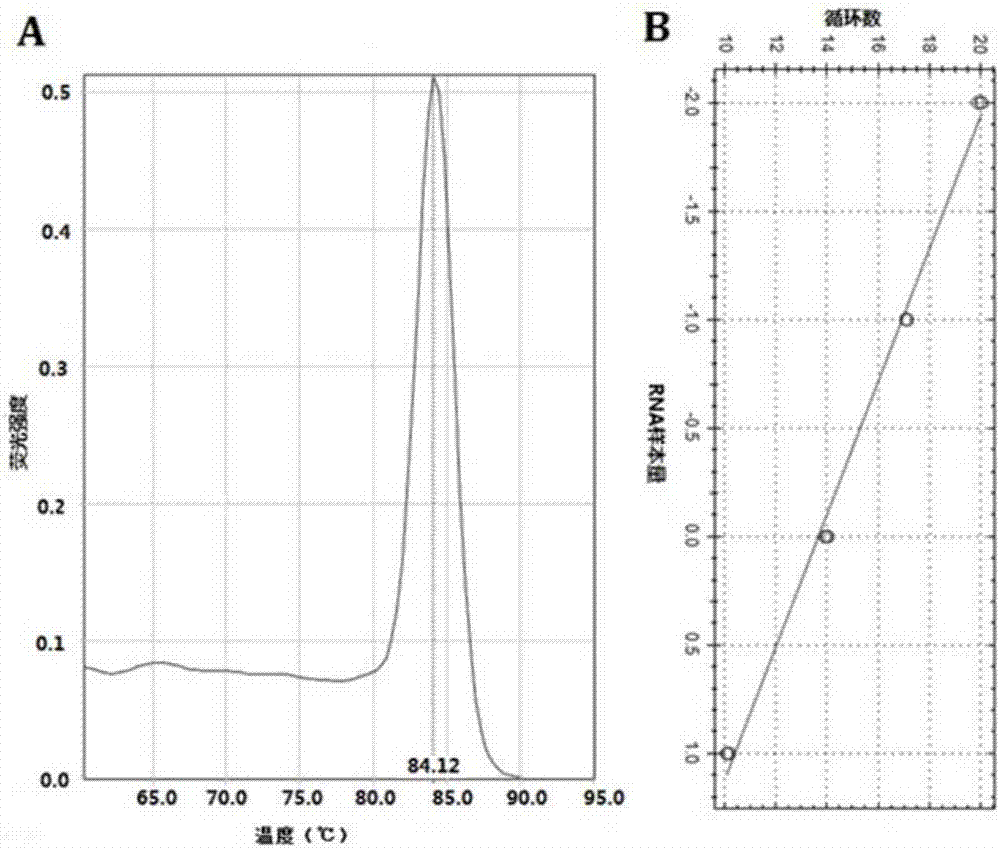
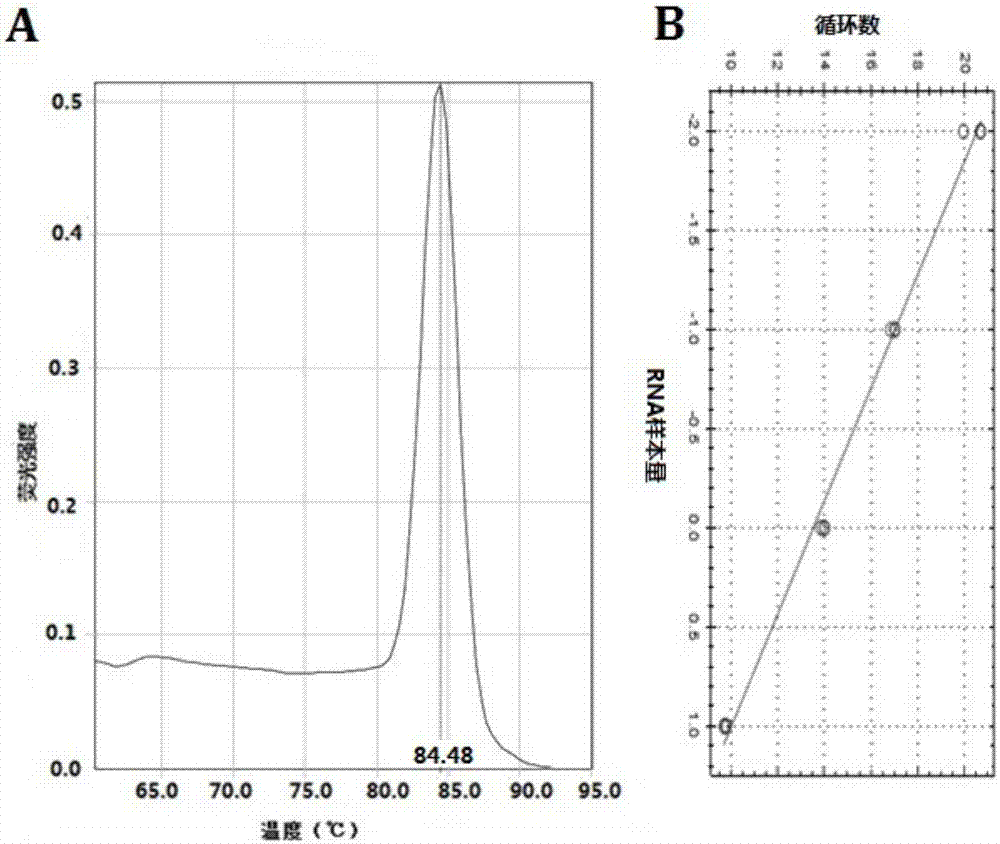
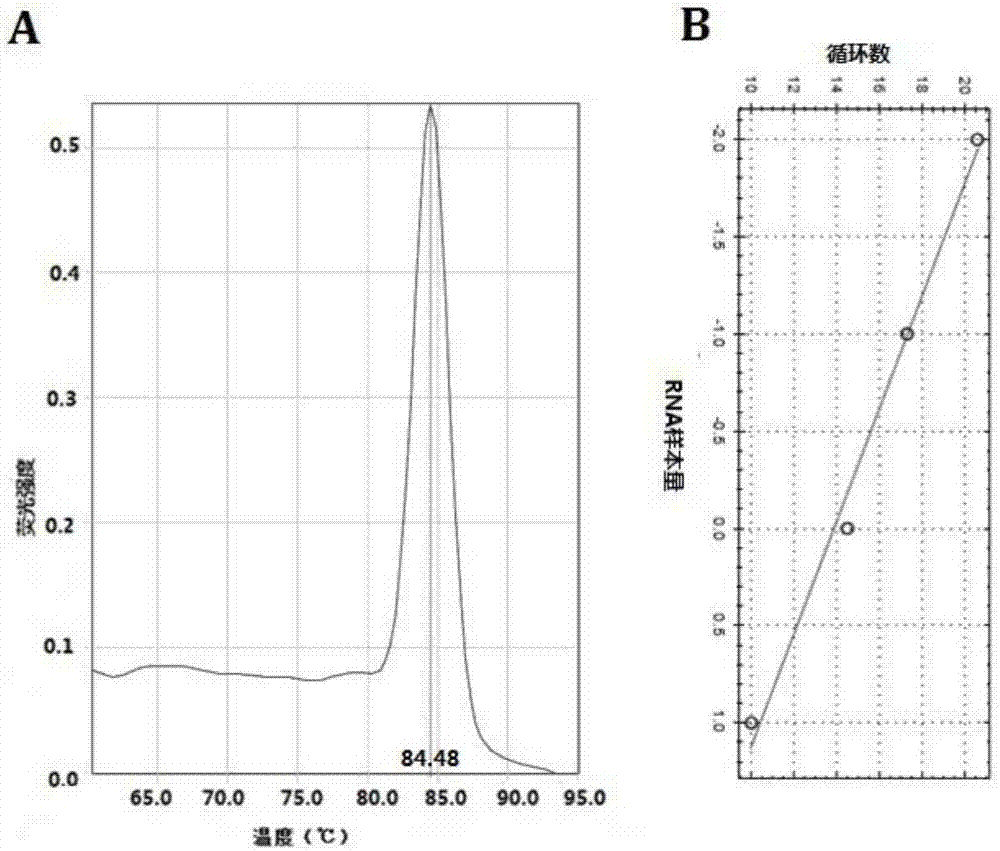
[0054] qRT-PCR program: 95°C 30s pre-denaturation; 95°C 3s, 30s, 60°C 30s, 40 cycles. Internal reference for U6 miRNA. Use ABI 7500fast Real-Time PCR instrument and software for analysis. RNA relative expression = 2 -△△CT , ΔCT=target gene CT-internal reference CT ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com