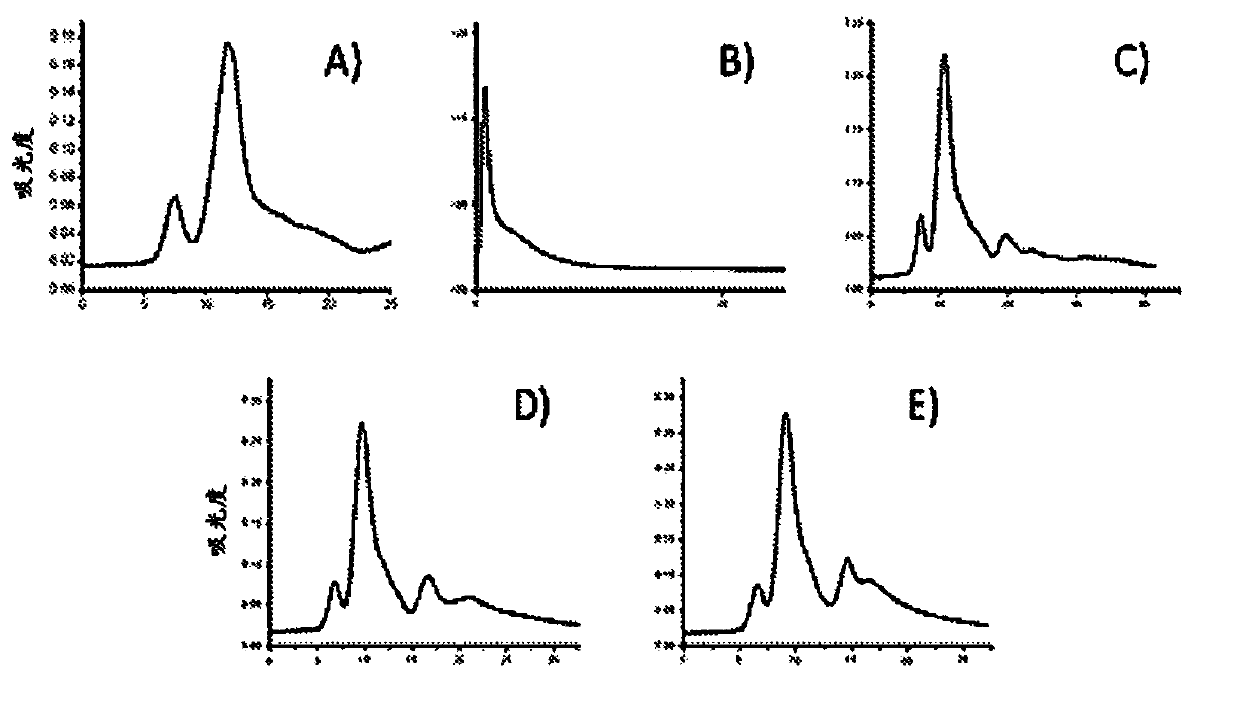
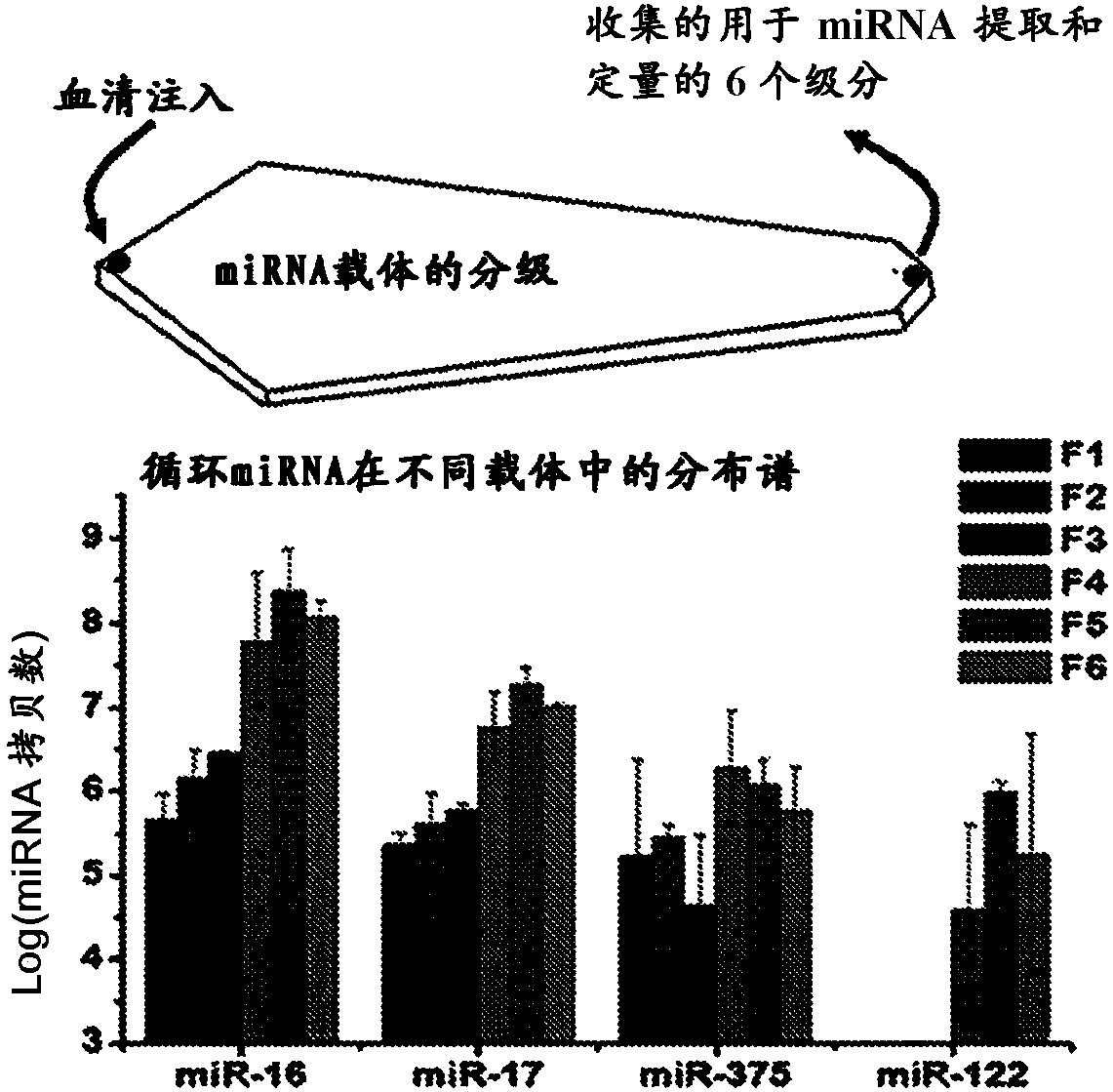
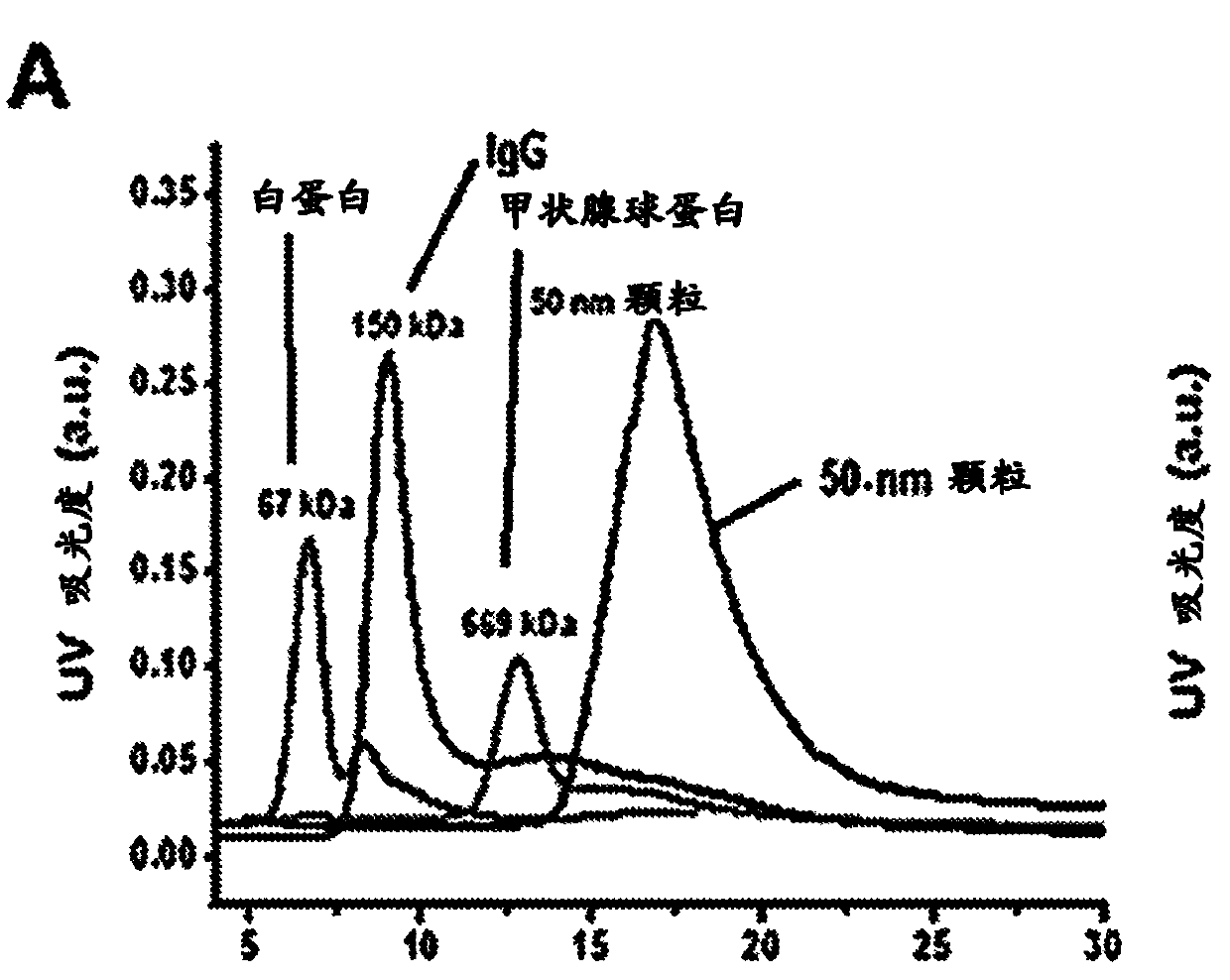
Methods to determine the distribution profiles of circulating rnas
A grading method and fractional technology, which can be applied in the fields of DNA/RNA fragments, recombinant DNA technology, biochemical equipment and methods, etc., and can solve problems such as unrevealed miRNA markers.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0084] Chemicals and biological materials. HDL and low density lipoprotein (LDL) were purchased from CalBioChem (EMD Millipore, Billerica, MA). Trizol LS reagent, 3,3'-Dioctadecyloxacarbonylcyanine periodate (DiO) and total exosome isolation kit were purchased from Invitrogen (Life Technologies). MicroRNA standards were purchased from Integrated DNA Technologies (Coralville, IA). TaqMan MicroRNA Assays specific for each miRNA strand were purchased from Applied Biosystems (Life Technologies). For the preparation of 1X PBS (10mM Phosphate, 137mM NaCl, 2.7mM KCl, and 1.0mM MgCl at pH 7.4 2 ), ethylene glycol, dimethyl sulfoxide, guanidine hydrochloride, RNA grade glycogen, 2-propanol, and chloroform were purchased from Thermo Fisher (Pittsburgh, PA). All single proteins used as AF4 standards were purchased from Sigma-Aldrich (St. Luis, MO). Taq 5X master mix was purchased from NewEngland Biolabs.
[0085] Serum samples. The serum samples used for exosome extraction and isolat...
Embodiment 2
[0121] Microchip manufacturing. Briefly, as Figure 13 Fabrication of microchips is generally shown in . A microchip platform was fabricated by bonding together a 3" x 1" glass slide (0.5 mm thick) and a cured PDMS substrate. To fabricate the PDMS substrates, a total of three hosts were prepared. First, a first body containing only channel features, Body-1, was fabricated by an open process from thiolene-based optical adhesive NOA81. In this method, by using a collimated UV light source (365nm, ~8.3mW / cm 2 ) for 5 seconds to pre-cure NOA81 between the glass slide (plasma-treated) and the PDMS bench. The thickness of Body-1 was determined by a spacer (~400 μm) placed between the glass slide and the PDMS stage, and the features were defined by a photomask. After a brief UV exposure, the slide was slowly removed from the PDMS stage with pre-cured NOA81 based channel features on the surface. Unexposed adhesive was removed using a syringe by sequential flushing with ethanol, a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com