Novel micro-environment biological macromolecule general oscillator as well as synthesis method and application thereof
A biomacromolecule and synthesis method technology, which is applied in the field of microenvironment biomacromolecule general oscillator and its synthesis, to achieve the effect of improving the scope of application and simplifying the research scheme
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0038] The synthesis method of the microenvironment biomacromolecule universal oscillator is as follows: using origami software, using a scaffold chain as a skeleton, and using a staple chain to fix the scaffold chain into the above-mentioned triple clip structure, which includes the following steps:
[0039] (1) First set the length, width and height of the base 4, then determine the position, length and number of double helix strands of each clip in the triple clip structure, and the specific position of the adapter 10 on each clip;
[0040] (2) adjust the position, length and the number of double helix strands of the staple chain, add the sequence of the scaffold chain, and automatically generate the sequence of all the staple chains;
[0041] (3) Experimental synthesis of the scaffold chain and the staple chain described in step (2).
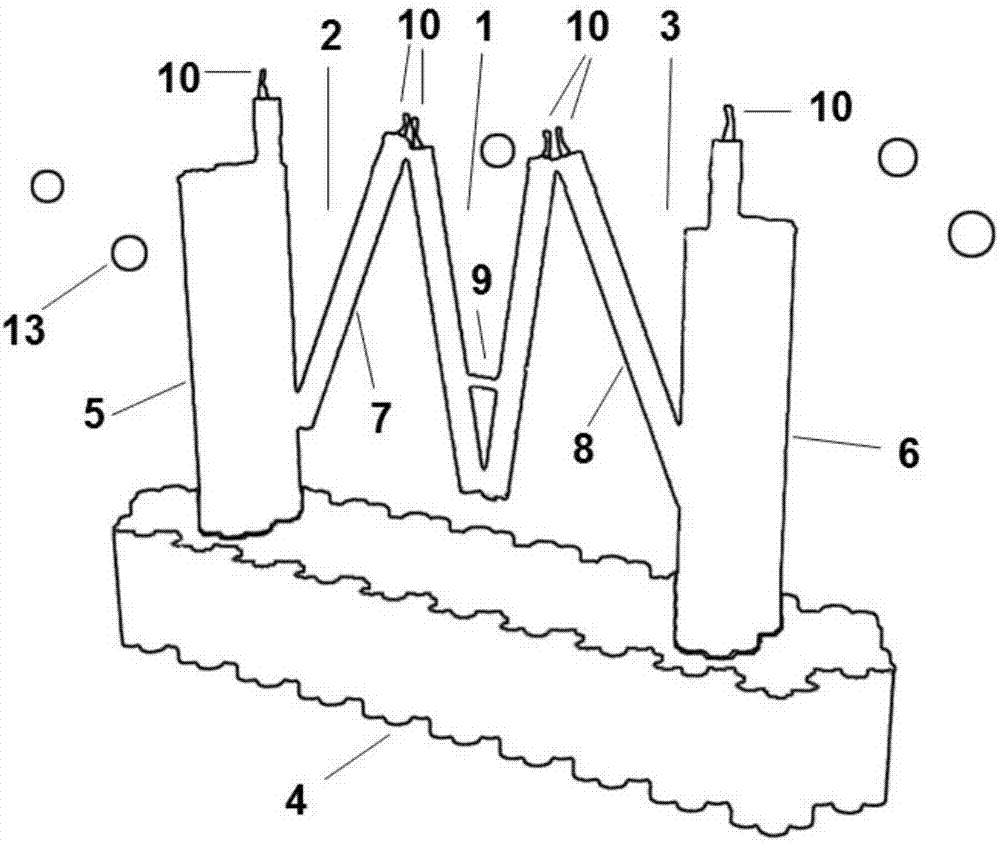
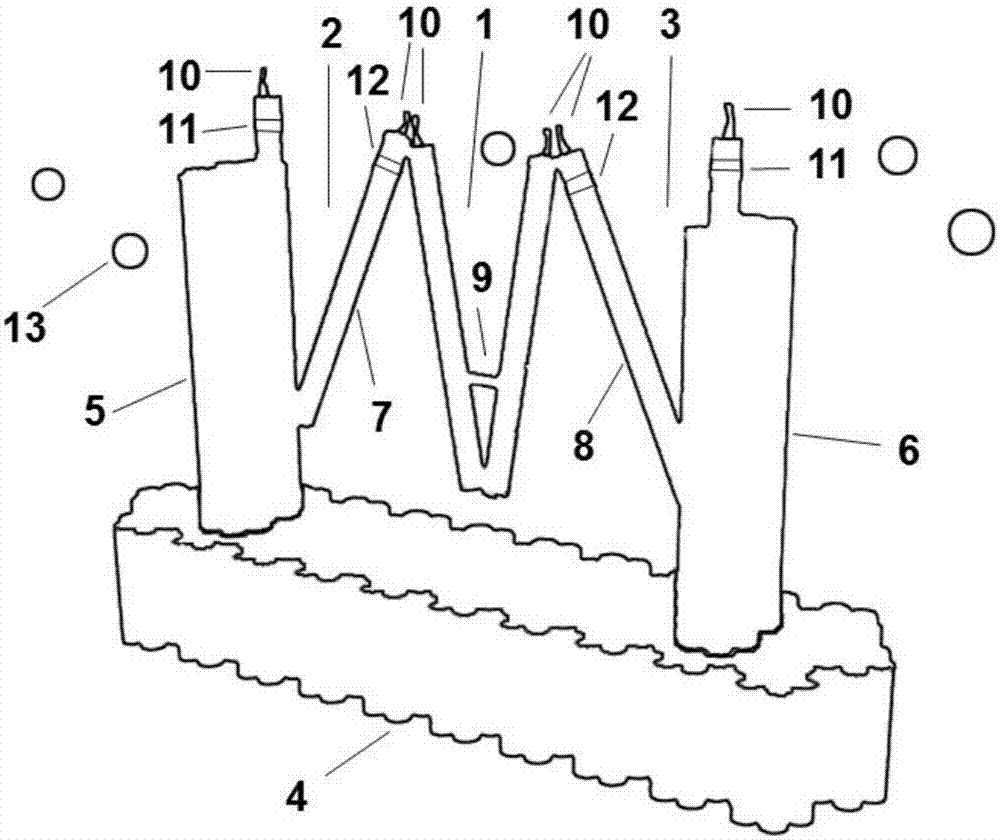
[0042] Wherein, step (1) first determines the positions of the first peripheral clip extremity 5 and the second peripheral clip extremity 6...
Embodiment 1
[0054] A new general-purpose oscillator for microenvironmental biomacromolecules, see attached figure 2 , its specific synthesis steps are:
[0055] Step 1: Use CADnano software to design the triple clip structure, determine the length, width, and height of the DNA structure base 4, and determine each clip in the triple clip structure (the first peripheral clip 2, the middle clip 1 and the second peripheral clip 3) The length and the number of double helix strands determine the specific position of the aptamer 10 (Aptamer) on each clip.
[0056] The specific content of the design of the triple clip structure is: the gene of the M13mp18 phage is the skeleton (scaffold chain), combined with the staple chain to form the DNA base 4 and the first peripheral clip outer limb 5 and the second peripheral clip outer limb 6, by The staple chain synthesizes the two limbs of the middle clip 1 and the inner limb 7 of the first peripheral clip and the inner limb 8 of the second peripheral ...
Embodiment 2
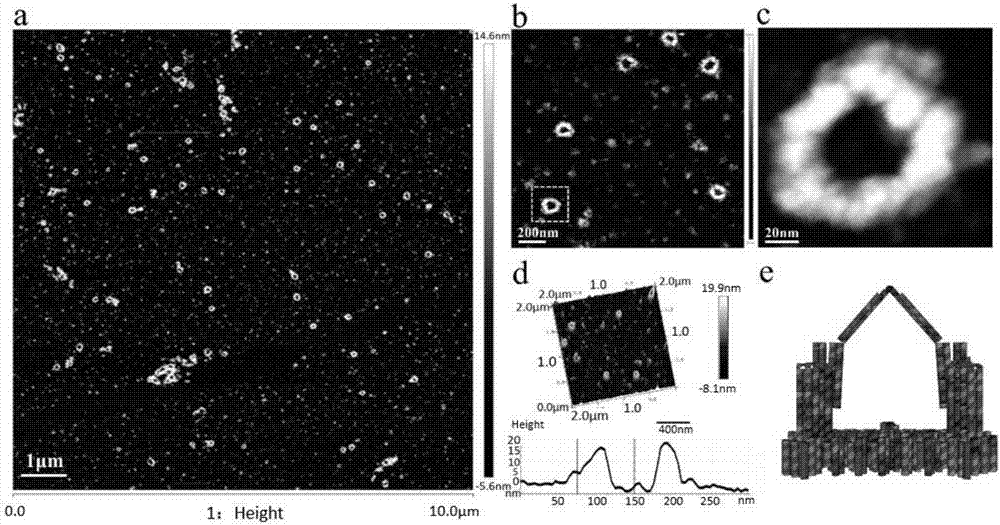
[0065] The oscillator prepared in Example 1 was observed using an atomic force microscope. The specific sample preparation and observation steps were as follows: first soak the cover glass in a PBS solution of 0.1 mg / mL poly-L-lysine overnight, and then Add 7uL sample to the surface of the coverslip and let it stand at room temperature for 10 minutes, then rinse the surface of the coverslip twice with 100uL 1xTAE solution, and keep it wet during the observation with the atomic force microscope.
[0066] Figure 4It is the triple clip structure (oscillator) of Example 1 observed under an atomic force microscope. a and b are the oscillators observed with the atomic force microscope under different magnifications; c: Atomic force microscope pictures when the middle clip 1 is opened and the first peripheral clip 2 and the second peripheral clip 3 are closed, because the middle clip 1 and the base The charges of seat 4 are the same and repel each other, causing the middle clip 1 t...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Spiral diameter | aaaaa | aaaaa |
| Longitudinal length | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com