A method for constructing a maternal plasma cell-free dna library and a method for typing paternal alleles
A DNA library and allele technology, applied in the field of biology, to achieve the effect of reducing cost and cycle, wide application range and simple method
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
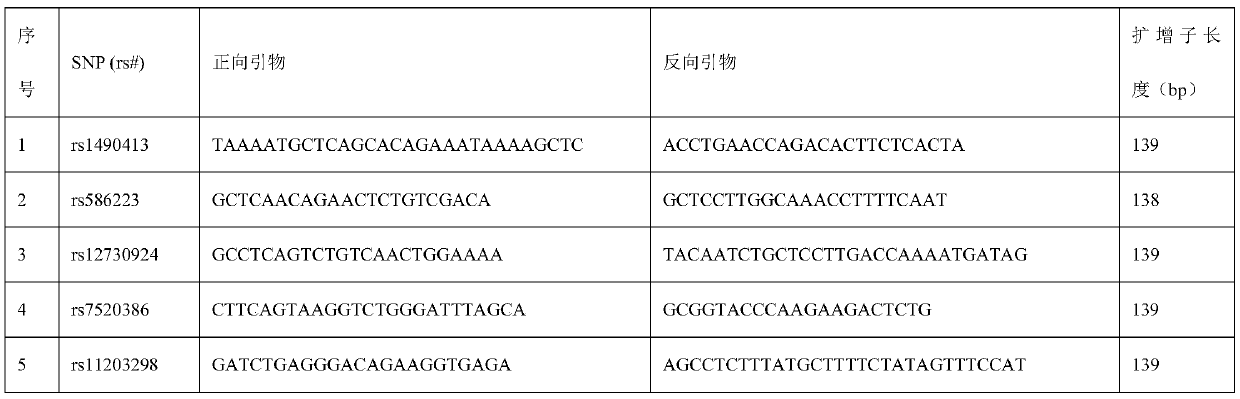
[0044] According to the gene sequences of 22 human autosomes and Y chromosomes, specific amplification primers were designed for 720 SNP sites evenly distributed in the genome (as shown in Table 1, and the sequences are shown in SEQ ID NO.1-1440 in turn. ), with a wide range of applications, and the amplicon length does not exceed 140bp.
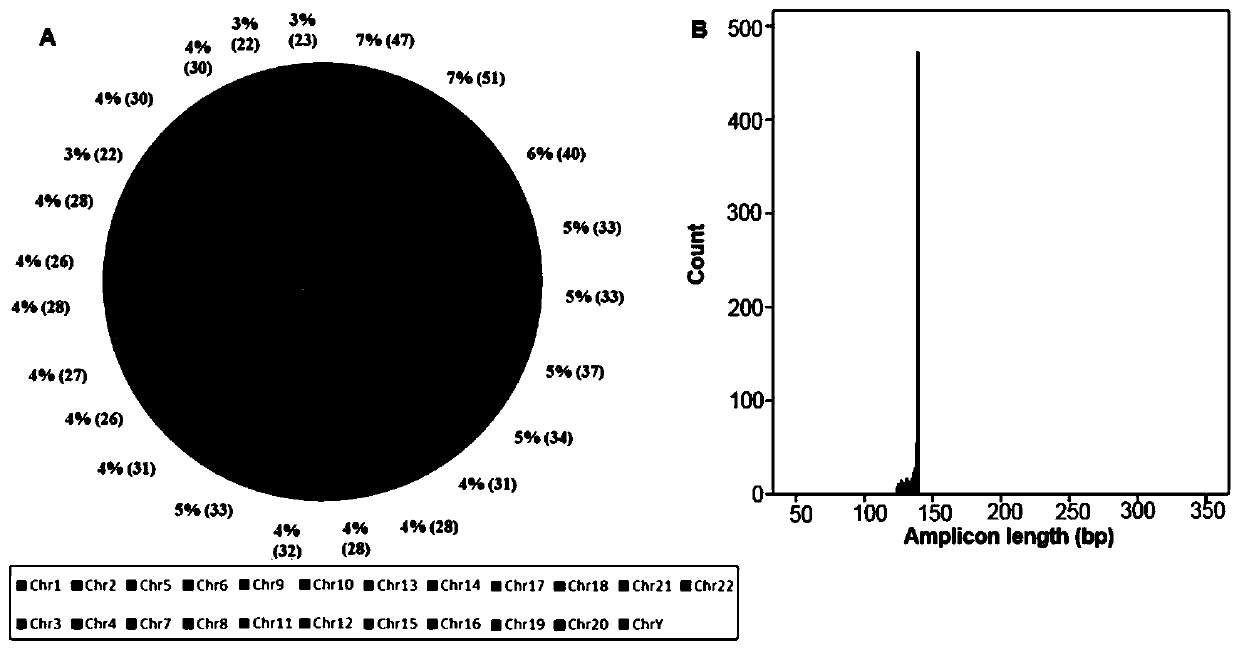
[0045] The distribution (A) of the 720 target SNPs detected by this primer set on each chromosome and the length distribution (B) of SNP amplicons are as follows figure 1 shown. 697 SNP sites (A-SNPs) located on 22 autosomes and 23 SNP sites (Y-SNPs) on the Y chromosome were selected and specific primers were designed. The lengths of the 720 amplicons were very concentrated, more than 80% of them were 136-139bp in length, the average was 137bp, the shortest was 124bp, and the longest was 139bp.
[0046] Table 1
[0047]
[0048]
[0049]
[0050]
[0051]
[0052]
[0053]
[0054]
[0055]
[0056]
[0057] ...
Embodiment 2
[0074] This implementation takes a pregnant woman at 12+5 weeks of pregnancy as an example to describe the content of the present invention in detail. The free DNA based on the high-throughput sequencing platform used in the present invention The method for DNA library construction, sequencing and paternal SNP allele detection comprises the following steps:
[0075] 1. Sample collection, processing and DNA extraction provide templates for library preparation in subsequent steps
[0076] Following the principle of "informed consent", 5 mL of peripheral blood was drawn from pregnant women before puncture, stored in EDTA anticoagulant tubes, centrifuged at 1600 g for 10 min at 4 °C within 8 hours, and the supernatant was centrifuged at 16000 g for 10 min to harvest plasma; the precipitated blood cell part was 10000 g Centrifuge for 5 minutes, and discard the residual supernatant. Fetal villus tissue was collected by B-ultrasound under sterile conditions and washed thoroughly wit...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com