Fluorescence sensation method for detecting exonuclease I
An exonuclease and fluorescence sensing technology is applied in the field of fluorescence biosensors to achieve accurate results, less reagent consumption and good selectivity.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] The technical solution adopted by the present invention to solve the above-mentioned technical problems is: a kind of fluorescence sensing method that detects Exo I, concrete steps are as follows:
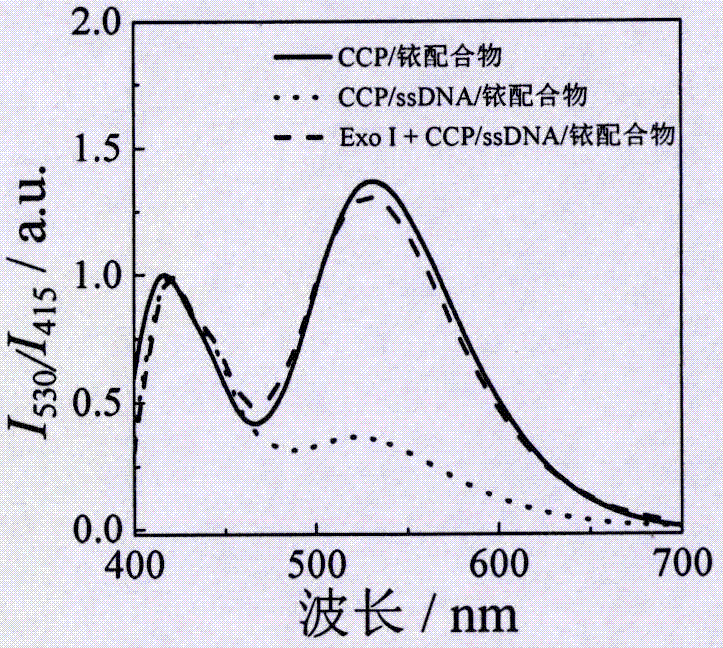
[0027] (1) Mix 90 μL of 3.28 μM CCP with 1 μL of 1 mM iridium complex, and then add a corresponding volume of 10×Exo I buffer to obtain 100 μL of fluorescence based on the FRET effect of CCP and iridium complexes for detection of Exo I sensor system. The fluorescence intensity was detected, and the ratio of the fluorescence intensity of the CCP (415nm) to the fluorescence intensity of the iridium complex (530nm) was calculated.
[0028] (2) Add 2 μL of 10 μM ssDNA to the system in step (1), detect the change of fluorescence intensity, and calculate the ratio of the fluorescence intensity of CCP (415nm) to the fluorescence intensity of the iridium complex (530nm).
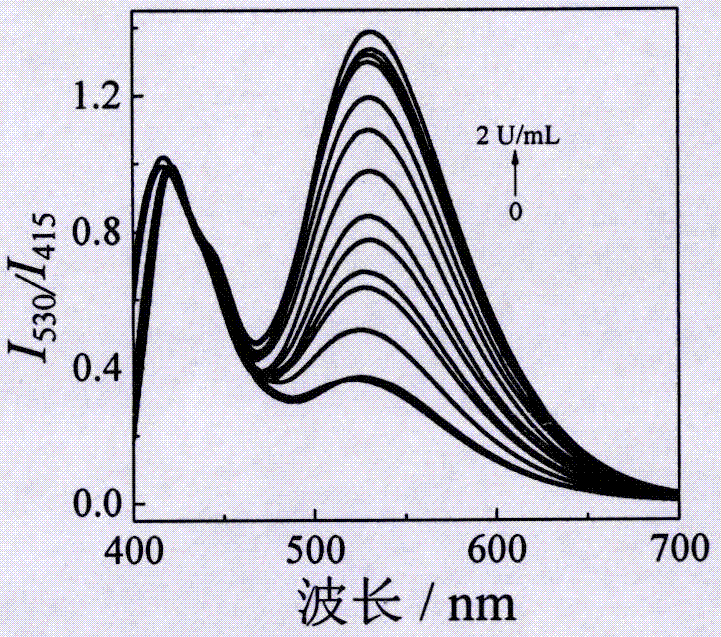
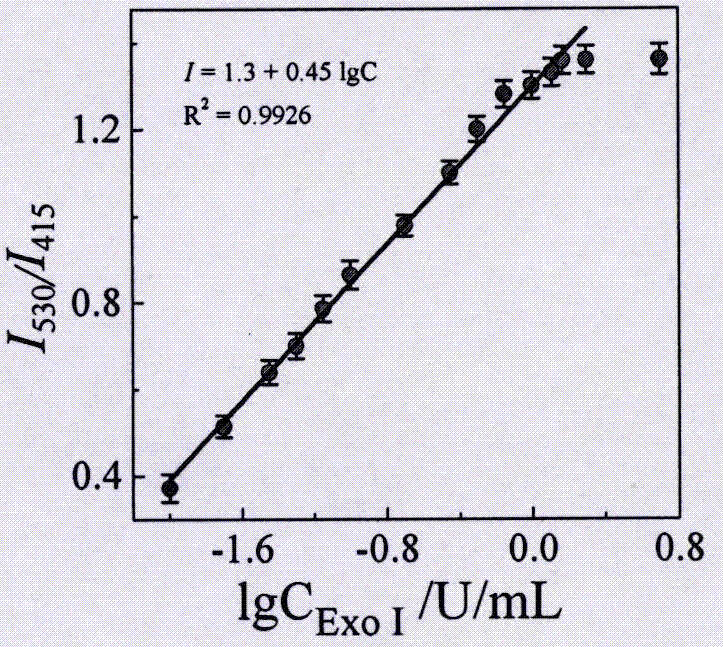
[0029] (3) Different concentrations of Exo I were added to step (2), and then the corresponding volume of 10×E...
Embodiment 2
[0031] The technical solution adopted by the present invention to solve the above-mentioned technical problems is: a kind of fluorescence sensing method that detects Exo I, concrete steps are as follows:
[0032] (1) Mix 85 μL of 4 μM CCP with 2 μL of 1 mM iridium complex, and then add a corresponding volume of 10× Exo I buffer to obtain 100 μL of a fluorescent sensor based on the FRET effect of CCP and iridium complexes for detecting Exo I system. The fluorescence intensity was detected, and the ratio of the fluorescence intensity of the CCP (415nm) to the fluorescence intensity of the iridium complex (530nm) was calculated.
[0033] (2) Add 1 μL of 15 μM ssDNA to the system in step (1), detect the change of fluorescence intensity, and calculate the ratio of the fluorescence intensity of CCP (415nm) to the fluorescence intensity of the iridium complex (530nm).
[0034](3) Different concentrations of Exo I were added to step (2), and then the corresponding volume of 10×Exo I ...
Embodiment 3
[0036] The technical solution adopted by the present invention to solve the above-mentioned technical problems is: a kind of fluorescence sensing method that detects Exo I, concrete steps are as follows:
[0037] (1) Mix 95 μL of 3 μM CCP with 0.5 μL of 2 mM iridium complex, and then add a corresponding volume of 10× Exo I buffer to obtain 100 μL of fluorescence based on the FRET effect of CCP and iridium complexes for detection of Exo I sensor system. The fluorescence intensity was detected, and the ratio of the fluorescence intensity of the CCP (415nm) to the fluorescence intensity of the iridium complex (530nm) was calculated.
[0038] (2) Add 3 μL of 5 μM ssDNA to the system in step (1), detect the change of fluorescence intensity, and calculate the ratio of the fluorescence intensity of CCP (415nm) to the fluorescence intensity of the iridium complex (530nm).
[0039] (3) Different concentrations of Exo I were added to step (2), and then the corresponding volume of 10×Ex...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Width | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com