Application of nucleic acid molecule
A nucleic acid molecule, nucleotide sequence technology, applied in the application field of nucleic acid molecules, can solve problems such as lack of research
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0081] Example 1 mmu-miR-146b-5p directly regulates the target gene HMGB1
[0082] ①Bioinformatics prediction
[0083] The binding site between mmu-miR-146b-5p and HMGB1 3'-UTR region:
[0084] According to the bioinformatics software miRWalk, DIANA-microT, miRanda, MiRSystem, miRDB, miRMap, miRNAsMap, Pictar, PITA, RNA22, RNAhybrid and Targetscan, the miRNAs interacting with the 3'-UTR region of the HMGB1 gene were predicted.
[0085] ② Plasmid construction
[0086] Target gene HMGB1 3'-UTR region sequence:
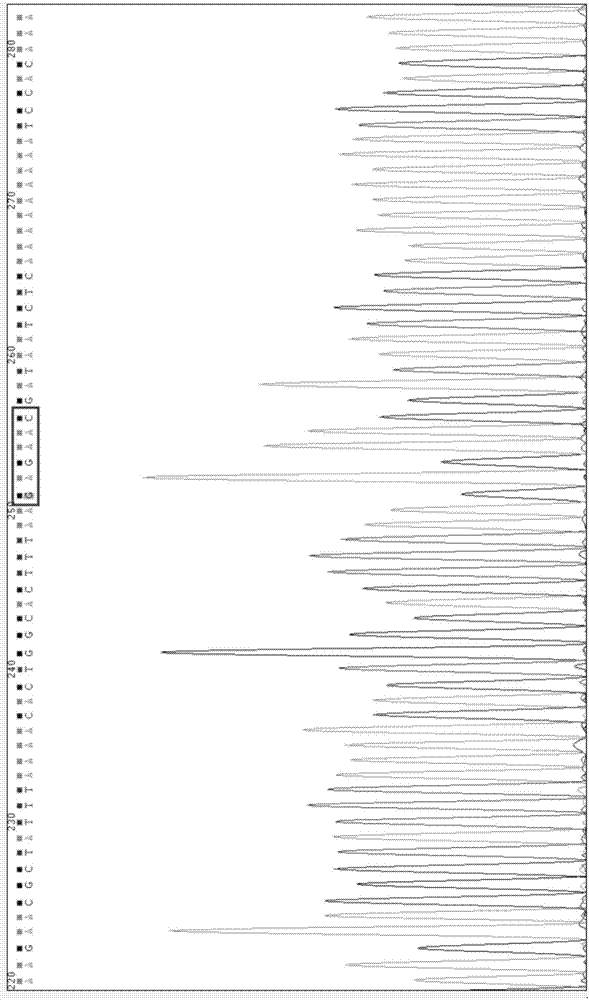
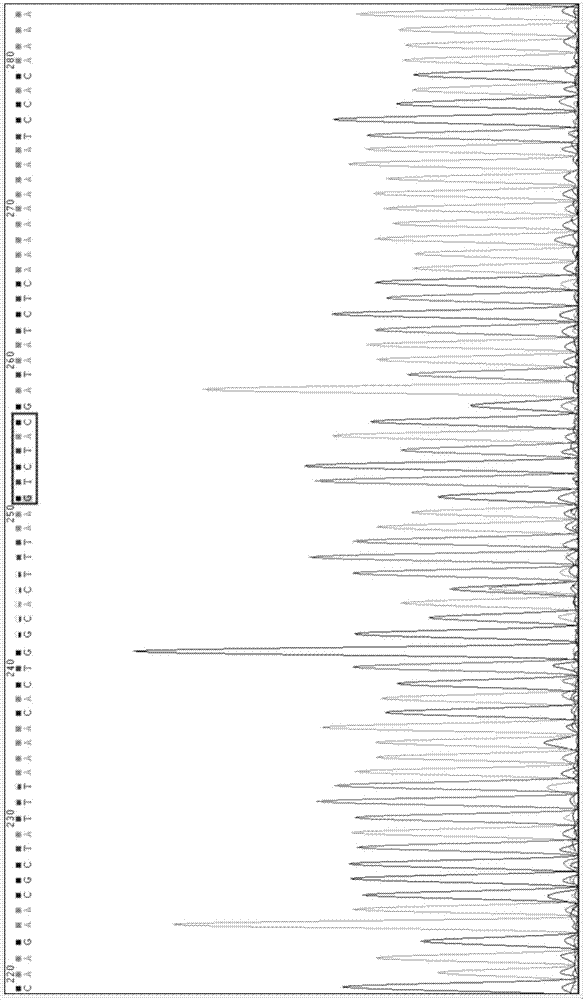
[0087] Through the bioinformatics database NCBI, the sequence of the mouse HMGB1 3'-UTR region containing the predicted binding site (gttctc, 1327-1332 nucleotides) was obtained, and cDNA was synthesized as a template, named wt-HMGB1 3'-UTR, The sequence is shown in SEQ ID No.13.
[0088] Point mutation HMGB1 3'-UTR region sequence:
[0089] The target gene HMGB1 3'-UTR region binding site (gttctc, 1327-1332 nucleotides) predicted by mmu-miR-146b-5p was subjected to...
Embodiment 2
[0135] Example 2 mmu-miR-146b-5p inhibits the expression level of the target gene HMGB1
[0136] ①Transfection of mmu-miR-146b-5p
[0137] To prepare OligoRNA:
[0138] OligoRNA sequences were purchased from Shanghai Yingwei Jieji Biotechnology Co., Ltd., and the sequences are shown in Table 3:
[0139] table 3
[0140]
[0141] Cell processing and grouping:
[0142] Candida albicans stimulated transfected macrophages with mimic NC, mmu-miR-146b-5p mimic, inhibitor NC and mmu-miR-146b-5p inhibitor for 36 hours, and divided into normal, C.albicans, mimic NC, mmu-miR-146b -5pmimic, inhibitor NC and mmu-miR-146b-5p inhibitor groups, collect samples for follow-up experiments.
[0143] Transfection of mmu-miR-146b-5p:
[0144](1) The extracted mouse primary peritoneal macrophages were divided into 3×10 6 Inoculate each well in a 6-well plate, add 2 mL of complete medium to each well, and place in an incubator (37°C, 5% CO 2 ) to incubate for 12h;
[0145] (2) Before tran...
Embodiment 3
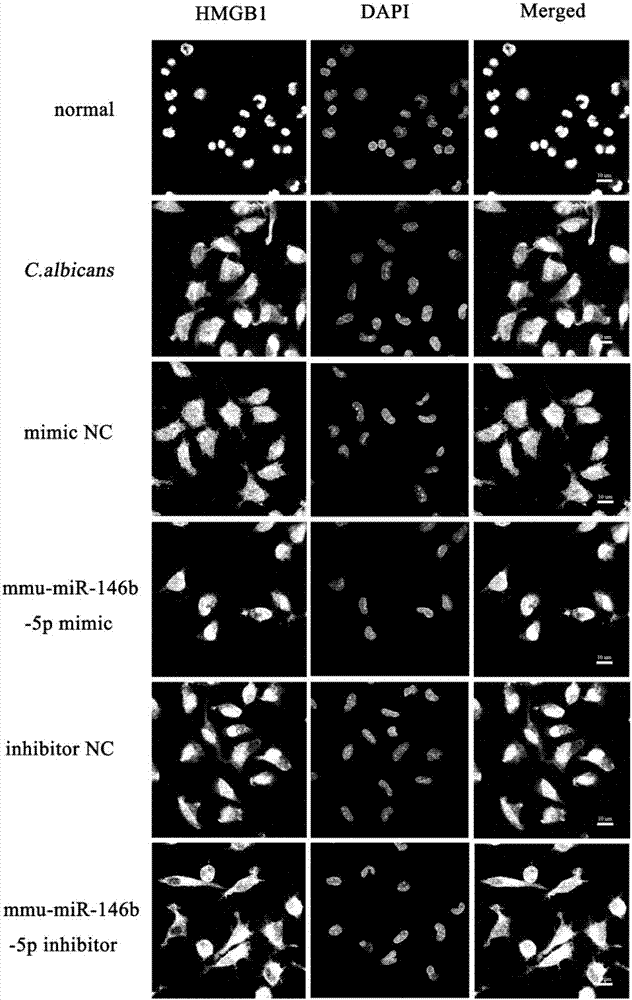
[0216] Example 3 mmu-miR-146b-5p can inhibit the translocation of HMGB1 in macrophages
[0217] Cell cytoplasmic protein and nuclear protein extraction:
[0218] According to the instructions of Jiangsu KGI Cytoplasmic and Nuclear Protein Extraction Kit, the BCA method was used for protein quantification, aliquoted and stored at -80°C.
[0219] Western blot experimental steps are as in Example 2.
[0220] Cell Laser Confocal Microscopy:
[0221] (1) Divide macrophages into 5×10 cells 5 Inoculate each well in a 24-well plate containing glass slides, add 1 mL of complete medium, and inoculate in an incubator (37°C, 5% CO 2 ) for 12 hours;
[0222] (2) OligoRNA was transfected into cells and incubated for 36 hours, and inactivated Candida albicans was stimulated for 36 hours;
[0223] (3) After the cell culture is over, wash with PBS solution (1min×3 times);
[0224] (4) Add 1mL 4% paraformaldehyde into each well, fix at room temperature for 30min;
[0225] (5) Washing wit...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com