Biosensor for detecting peptidylarginine deiminases, and preparation method and application thereof
A peptidyl arginine deiminase and biosensor technology, which can be applied to instruments, measuring devices, scientific instruments, etc., can solve the problems of low detection sensitivity, difficult operation and complicated operation, and easy to be interfered by the external environment. Sensitive, efficient, selective, easy to operate, and highly selective
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] Example 1: Reaction under different modification conditions
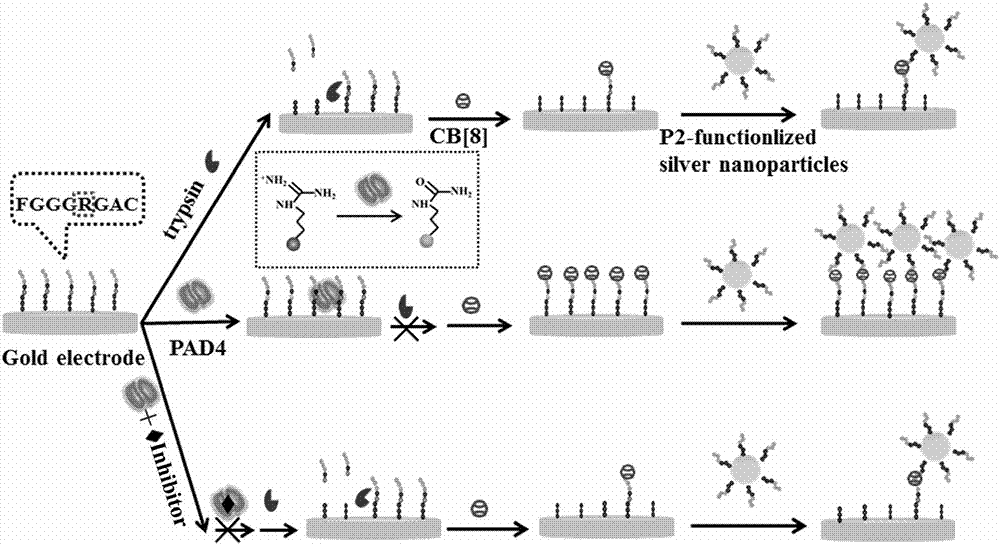
[0030] First, gold electrodes were polished with 1 μm, 0.3 μm and 0.05 μm aluminum powder. Then, the gold electrode was sonicated in ethanol and double-distilled water for 5 minutes each. Then the piranha (H 2 SO 4 :H 2 o 2 =3︰1) Purify for 5 minutes, rinse with double distilled water and use 0.5 M H2 SO 4 Electrochemical purification to remove any remaining impurities. After drying with nitrogen, the gold electrode was used to modify the polypeptide chain 1. React at 4°C for 16-18h; each 100μL modified reaction solution consisted of 50μL, 10μM polypeptide 1, 30μL, 10mM PBS and 20μL, 50mM TCEP, and mixed them evenly. The electrodes are submerged in the solution. At this time, polypeptide chain 2 is also activated and modified with silver nanoparticles. 4 Reaction 16-18h. 60 μL of 5 μM polypeptide chain 2 was added to 1440 μL of the prepared silver nanoparticle solution.
[0031] Cucurbituril 8 can c...
Embodiment 2
[0032] Example 2: Detection of different concentrations of PAD4
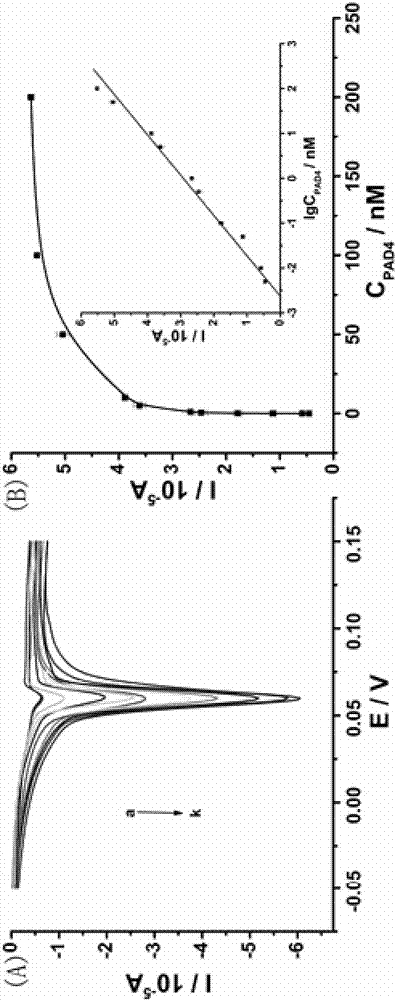
[0033] Add different concentrations of PAD4 to the reaction system (0.005nM, 0.01nM, 0.05nM, 0.1nM, 0.5nM, 1nM, 5nM, 10nM, 50nM, 100nM, 200nM ) The results are as follows image 3 shown. The activity of PAD4 to catalyze the citrullination of arginine increases with the increase of its concentration, and the peak current value also increases. image 3 The inset plot further illustrates the linearity of PAD4 from 0.005 nM to 200 nM. The linear equation is I (10 -5 A)=2.94233+1.12126lgC PAD4 (nM), R 2 =0.993. A detection limit of 0.79 pM was obtained at three times the noise ratio.
Embodiment 3
[0034] Example 3: Research on the Effect of PAD4 Inhibitors
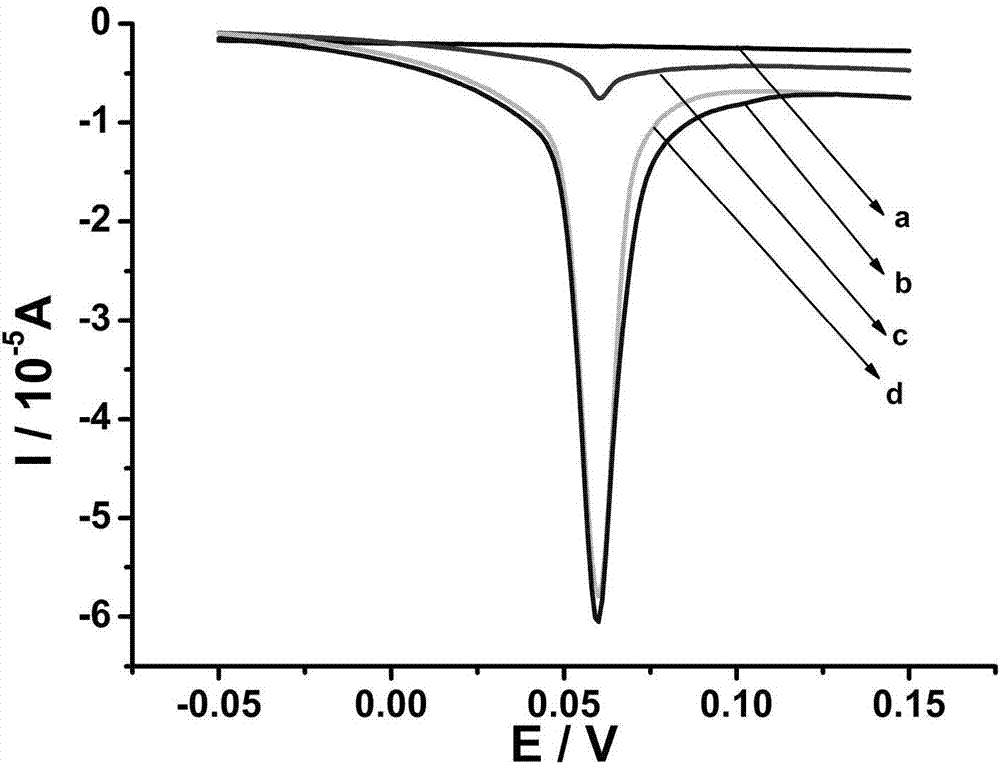
[0035] For the research on PAD4 inhibitors, it is generally the research on the mechanism of action of chloramidine on its inhibitors. Chloramidine is an irreversible PAD4 inactivator, and the combination of the inhibitor will destroy the structure of the active site of PAD4, thereby greatly reducing the activity of PAD4. Figure 4 The effect of different concentrations of inhibitors on PAD4 activity is shown. The activity of PAD4 was greatly reduced with increasing inhibitor concentration. The inset plots represent the concentration of the inhibitor chloramidine versus the rate of inhibition.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com