Jerusalem artichoke fructan exo-hydrolase gene 1-FEH III and protein encoded by same and application
A technology of 1-FEHIII and fructan, applied in the fields of hydrolase, application, genetic engineering, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] Cloning of embodiment 1 Jerusalem artichoke Ht1-FEH III
[0029] With upstream primer primer ATGATGAAGATTTATGGGTTTTG (SEQ ID NO.3) and downstream primer TTATATAATAGGCTTTCTTC (SEQ ID NO.4) and Jerusalem artichoke tuber cDNA (mRNA reverse transcription) as template and common Taq enzyme (CW Kangwei Company) for PCR amplification , the reaction system is as follows:
[0030]
[0031] The amplified product was run on the gel and photographed on the gel, and the band of the corresponding size was cut out for recovery. After connecting with the carrier PMd-19, it was transferred to Escherichia coli and sent for sequencing. After the sequencing was correct, the plasmid was extracted with a plasmid extraction kit (CW Kangwei Co., Ltd.). All operating steps refer to Instructions are carried out.
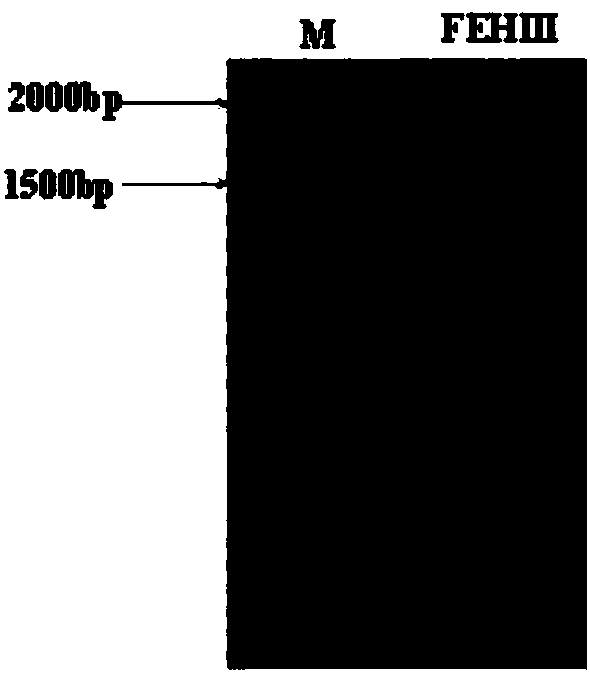
[0032] According to the prediction of the Jerusalem artichoke database, the size of the spliced Ht1-FEH III gene is 1719bp. Using cDNA as a template and using specific primers f...
Embodiment 2
[0035] 2.1 Construction of expression vector
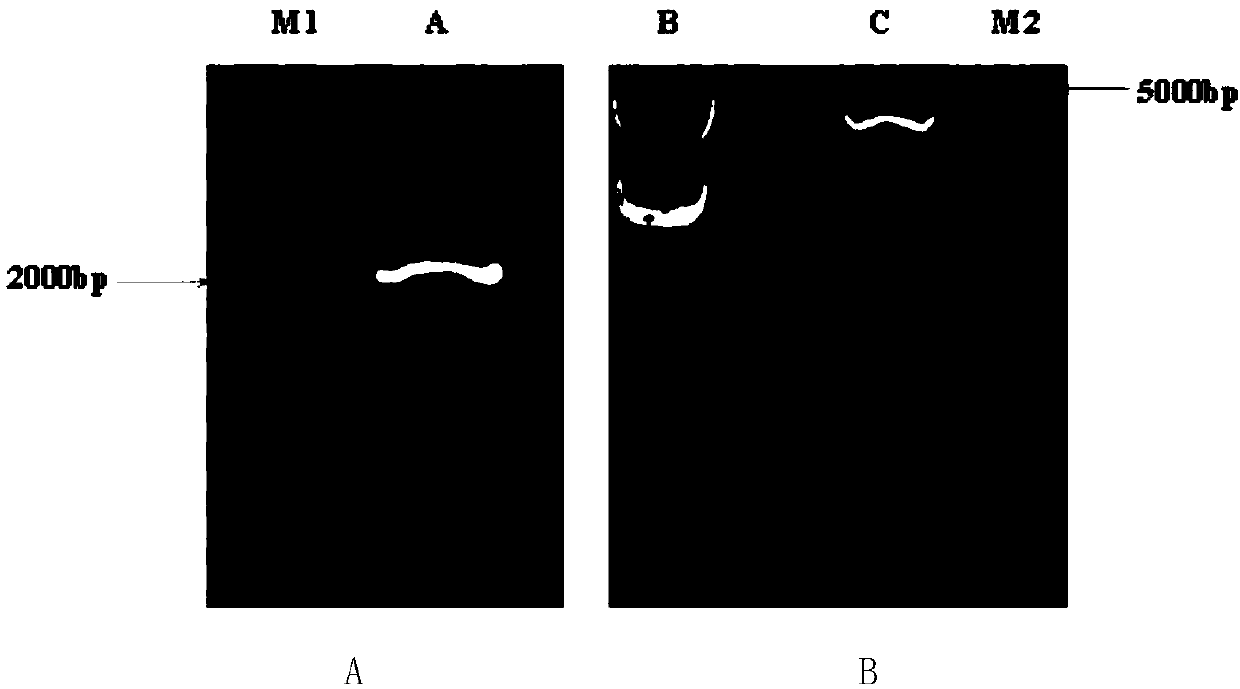
[0036] Using pPiczαC as a vector, because it has a 6xHIS tag, it can be expressed in Pichia pastoris and attached with affinity Ni in the purification stage + column to achieve the purpose of purification. Ht1-FEH III was amplified by high-fidelity enzyme (TaKaRa), and PCR amplification was performed with the above cloned Ht1-FEH III, and restriction sites XhoI / NotI were added to both ends of the gene ( image 3 Left picture), connect the pEASY-Blunt (TransGen) vector for transformation, select positive clones for sequencing, and extract the plasmid for double enzyme digestion after the sequencing is correct ( image 3 Right picture), in order to connect the target gene to the pPiczαC vector, pPiczαC was also digested with XhoI / NotI double enzymes, after digestion, it was ligated with T4 ligase (TaKaRa, D2011A), and after transfection into E. coli, it was placed on a low-salt LB plate containing Zeocin Positives in the screening...
Embodiment 3
[0043] Purification and SDS-PAGE of embodiment 3 recombinant protein
[0044] 3.1 Protein purification
[0045] After 5 days of induction, use a 4°C refrigerated centrifuge to remove the bacterial cell pellet and take the supernatant to obtain a crude enzyme solution, which is stored at -80°C. The purification process starts with 80% ammonium sulfate precipitation, 100ml of crude enzyme solution needs to be salted out by adding 51.6g of ammonium sulfate; pour 100ml of crude enzyme solution into a beaker placed on ice, and slowly stir while adding ammonium sulfate solid until Dissolve it completely, then put it in the refrigerator for salting out overnight, take it out and centrifuge the next day, and the protein precipitate obtained by centrifugation also needs to be washed twice with 80% ammonium sulfate solution and then centrifuged, and finally dissolved in 50mM acetic acid sodium acetate buffer solution with pH5.2 middle.
[0046] In order to further remove salt ions, th...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Protein molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com