Recombinant-plectasin-expressing Pichia pastoris
A technology of mycelia and Pichia pastoris, which is applied in the fields of biotechnology and genetic engineering, can solve the problems of reduced gene copy number of bacterial strains, heavy screening workload, unstable strains, etc., and achieves the goal of improving stability and reducing copy number Lost, increased dispersion effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0067] Example 1 Construction of Plectasin Expression Vector
[0068] 1.1 Design of plectasin NZ2114 gene based on the codon preference of Pichia pastoris
[0069] According to Pichia pastoris codon preference
[0070] Codon Frequency ‰ Quantity
[0071] UUU 24.1 1963
[0072] UCU 24.4 1983
[0073] UAU 16.0 1300
[0074] UGU 7.7 626
[0075] UUC 20.6 1675
[0076] UCC 16.5 1344
[0077] UAC 18.1 1473
[0078] UGC 4.4 356
[0079] UUA 15.6 1265
[0080] UCA 15.2 1234
[0081] UAA 0.8 69
[0082] UGA 0.3 27
[0083] UUG 31.5 2562
[0084] UCG 7.4 598
[0085] UAG 0.5 40
[0086] UGG 10.3 834
[0087] CUU 15.9 1289
[0088] CCU 15.8 1282
[0089] CAU 11.8 960
[0090] CGU 6.9 564
[0091] CUC 7.6 620
[0092] CCC 6.8 553
[0093] CAC 9.1 737
[0094] CGC 2.2 175
[0095] CUA 10.7 873
[0096] CCA 18.9 1540
[0097] CAA 25.4 2069
[0098] CGA 4.2 340
[0099] CUG 14.9 1215
[0100] CCG 3.9 320
[0101] CAG 16.3 1323
[0102] CGG 1.9 158
[0103] AU...
Embodiment 2
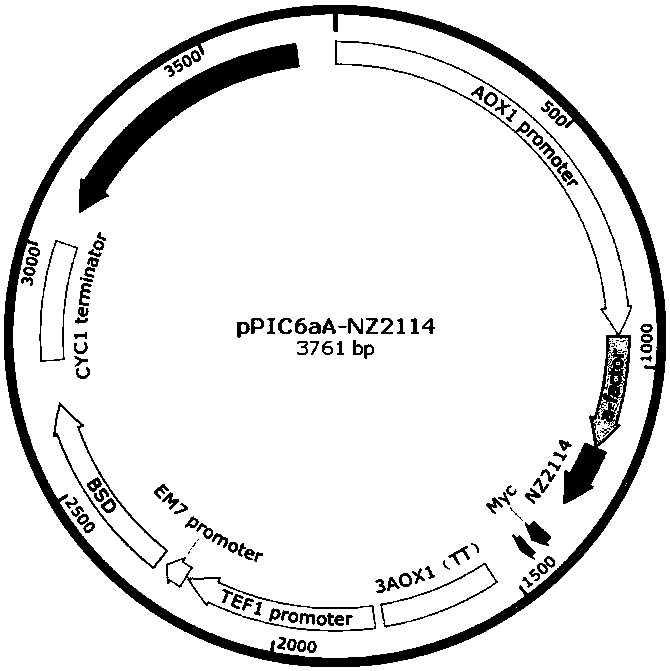
[0166] Embodiment 2 Construction of Plectasin Engineering Bacteria
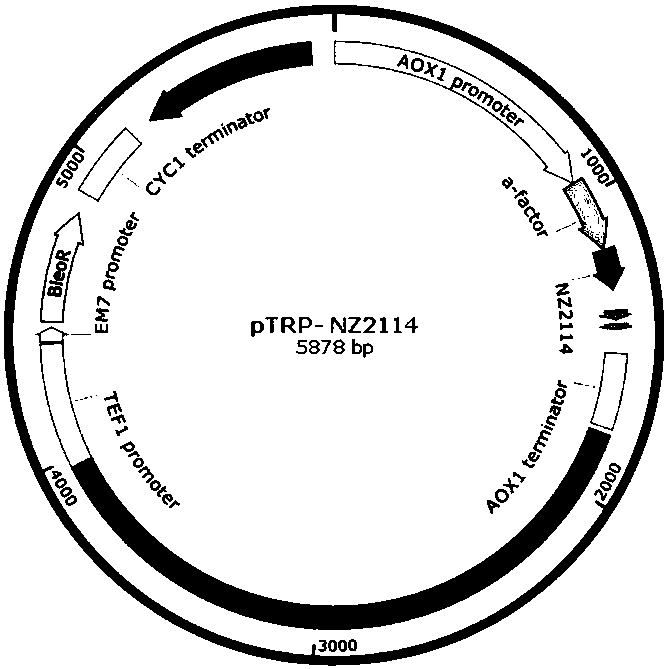
[0167] see Figure 6 , SpeI restriction endonuclease single-digestion treatment of pTRP-NZ2114 expression vector, cutting gel to recover linear expression vector, electroporation (1.5-2.5kv) to X33 Pichia competent cells or other Pichia such as GS115, SMD1168, etc. , spread the bacterial solution on a YPD plate (containing 0.25-4mg / mL bleomycin), and incubate at 30°C for 2-3 days until a single colony grows. Pick a single colony, cultivate and evaluate the expression of plectasin, and measure the size of the inhibition zone after the fermentation supernatant is diluted by a certain number of times. The optimal strain PLE-1 is selected for the next experiment.
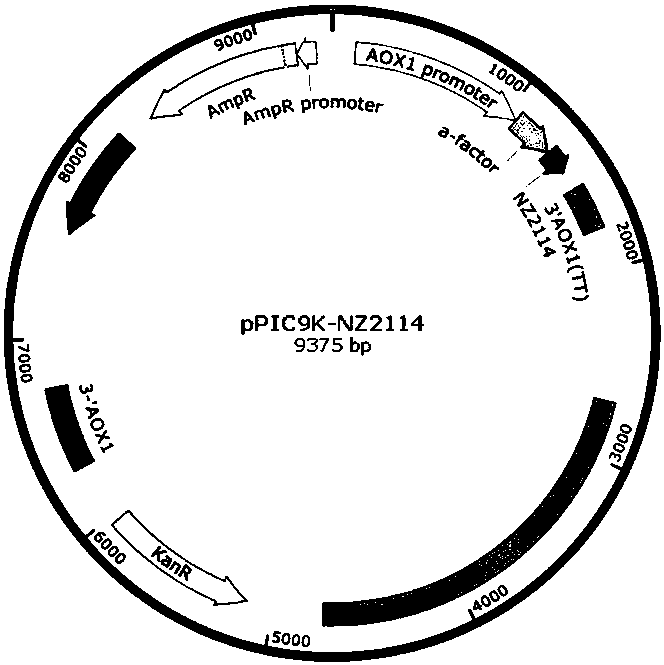
[0168] Such as Figure 7As shown, the pPIC9K-NZ2114 expression vector was treated with SalI restriction endonuclease single enzyme digestion, the linearized expression vector was recovered by cutting the gel, electroporated (1.5-2.5kv) to PLE-1 compe...
Embodiment 3
[0170] Example 3 Induced Expression of Recombinant Plectasin
[0171] The Pichia cells expressing Pichia pastoris prepared in Example 2 were picked, inoculated in 25 mL of BMGY medium, and cultured at 30° C. and 220 rpm for 24 hours to prepare a primary seed solution.
[0172] Inoculate 20 mL of primary seed liquid into 200 mL of BMGY medium to prepare secondary seed liquid, and culture at 30°C and 220 rpm for 24 hours.
[0173] All the secondary seed liquids were inoculated in a 5L fermenter (2L BSM medium), the temperature was controlled at 30°C±0.5°C, the dissolved oxygen was 20%±5%, and the pH=5.0±0.5.
[0174] After the basic glycerol was exhausted, 10% glycerol was added continuously. After the glycerol was exhausted, until the dissolved oxygen rose to 90%, starvation was performed for half an hour, and methanol was added to induce the expression of Plectasin. The total induction was 72 hours, and centrifuged (6000×g, 5min) to take The fermentation supernatant was teste...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com