Synthetic single guide RNA for Cas9-mediated gene editing
A single-guide and composition technology, applied in the field of gene editing, can solve the problems of low yield and the impossibility of practical application of chemical synthesis of single-guide RNA molecules
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
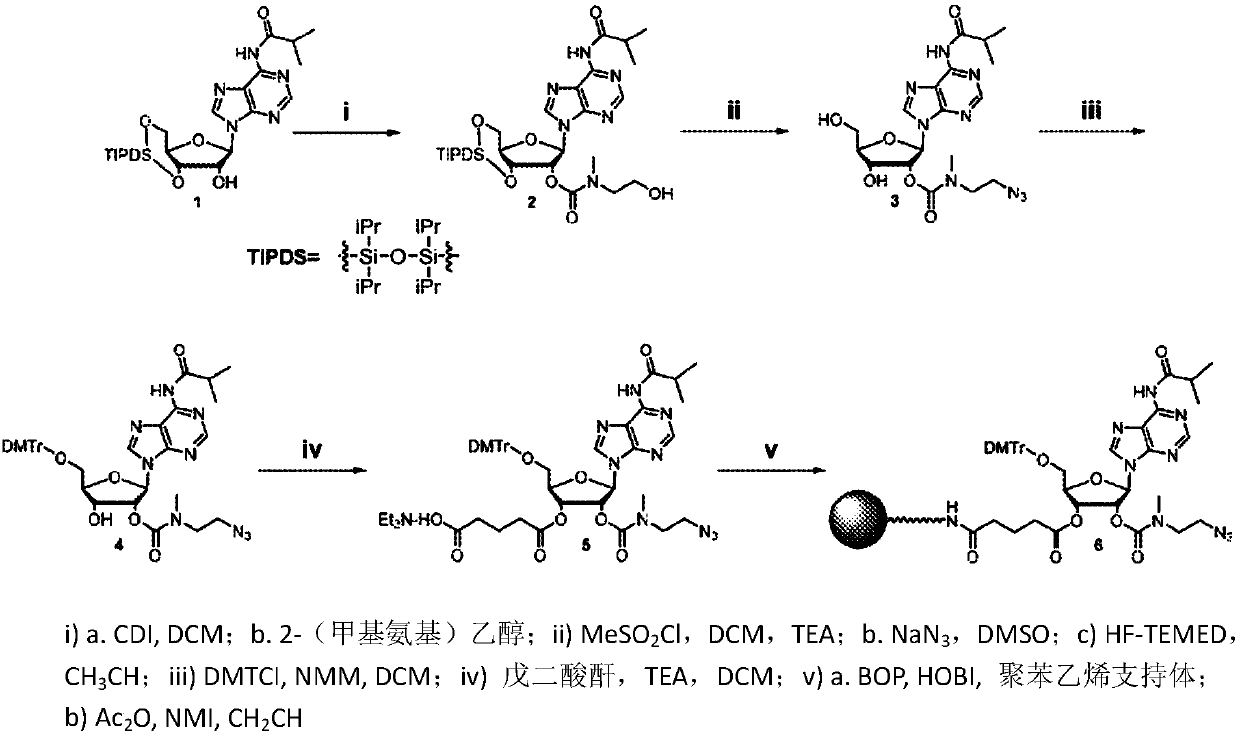
[0077] Example 1. Preparation of 3'-azidoadenosine polystyrene support ( figure 1 )
[0078] 1.
[0079] N 6 -Isobutyryl-2'-O-[2-(2-hydroxyethyl)methylcarbamate]-3',5'-O-(tetraisopropyl-di Siloxane-1,3-diyl)adenosine (2):
[0080] To a solution of compound 1 (10.0 g, 17.2 mmol) in 170 mL of dichloromethane (DCM) was added CDI (1,1'-carbonyldiimidazole) (2.9 g, 18.1 mmol). After stirring for 18 hours, 2-(methylamino)ethanol (5.2 g, 68.8 mmol) was added. After 1.5 hours the reaction was stopped and evaporated to dryness. The crude material was purified on a Biotage Isolera using a 100 g Ultra column with an ethyl acetate:MeOH gradient (0→10%) to afford 2 (10.8 g, 93%) as a white foam. Compound 2 was analyzed by RP-HPLC: 10.54 min, 99.4%. 1 H NMR (CDCl 3 , 300MHz) δ8.65(s, 1H), 8.63(s, 1H), 8.10(s, 1H), 6.04(d, J=8.8Hz, 1H), 5.64(d, J=5.3Hz, 1H), 5.15 (m, 1H), 4.16-3.98 (m, 4H), 3.76 (m, 2H), 3.56-3.15 (m, 3H), 3.05 and 2.96 (s, 3H each), 2.86 (s, 1H), 2.59 (m, 1H), ...
Embodiment 2
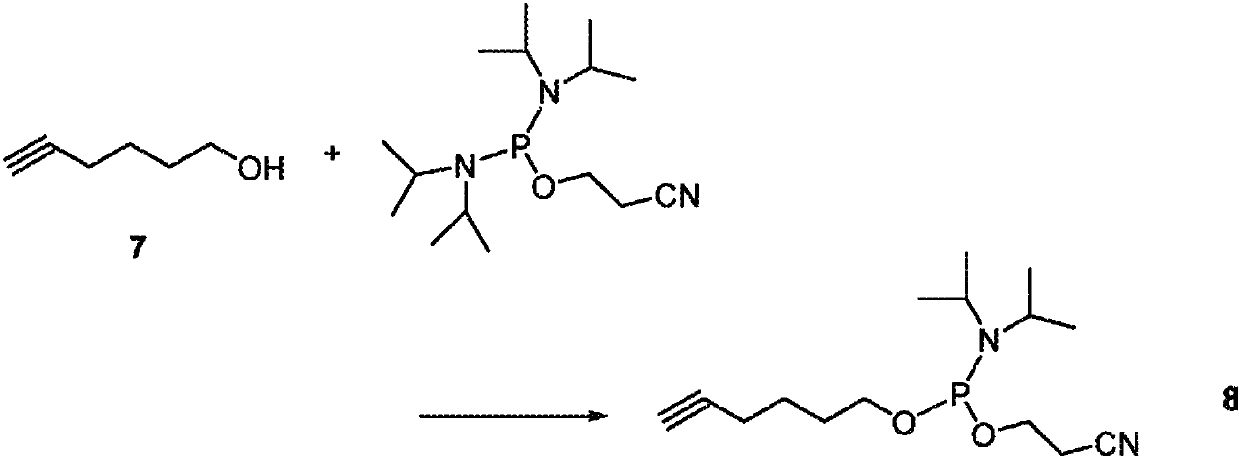
[0091] Embodiment 2. prepare 5'-hexyne phosphoramidite (8) ( figure 2 )
[0092] Compound 7 (hex-5-yn-1-ol, 1.4 mL) was dissolved in 10 mL of DCM in a flask, and N,N-diisopropylamine (1.82 mL) was added to the solution. In another flask under anhydrous conditions, dilute the phosphitylation reagent bis-(N,N-diisopropylamino)-cyanoethylphosphine (1.5 eq / eq 7), MeCN containing 0.45M 1H-tetrazole (0.5 eq tetrazole / eq 7) was added and shaken for 5 min. Next, the solution of the activated phosphitylation reagent was added to the well-stirred solution of compound 7 at room temperature, and stirred at room temperature until the reaction was complete by TLC analysis. To quench excess phosphine, ethanol was added and the reaction mixture was stirred for a further 30 min and dried on a rotary evaporator. The product was purified on silica gel to afford 0.8 g of phosphoramidite 8. 31 P NMR (CDCl 3 , 121.5MHz) δ147.0(s).
Embodiment 3
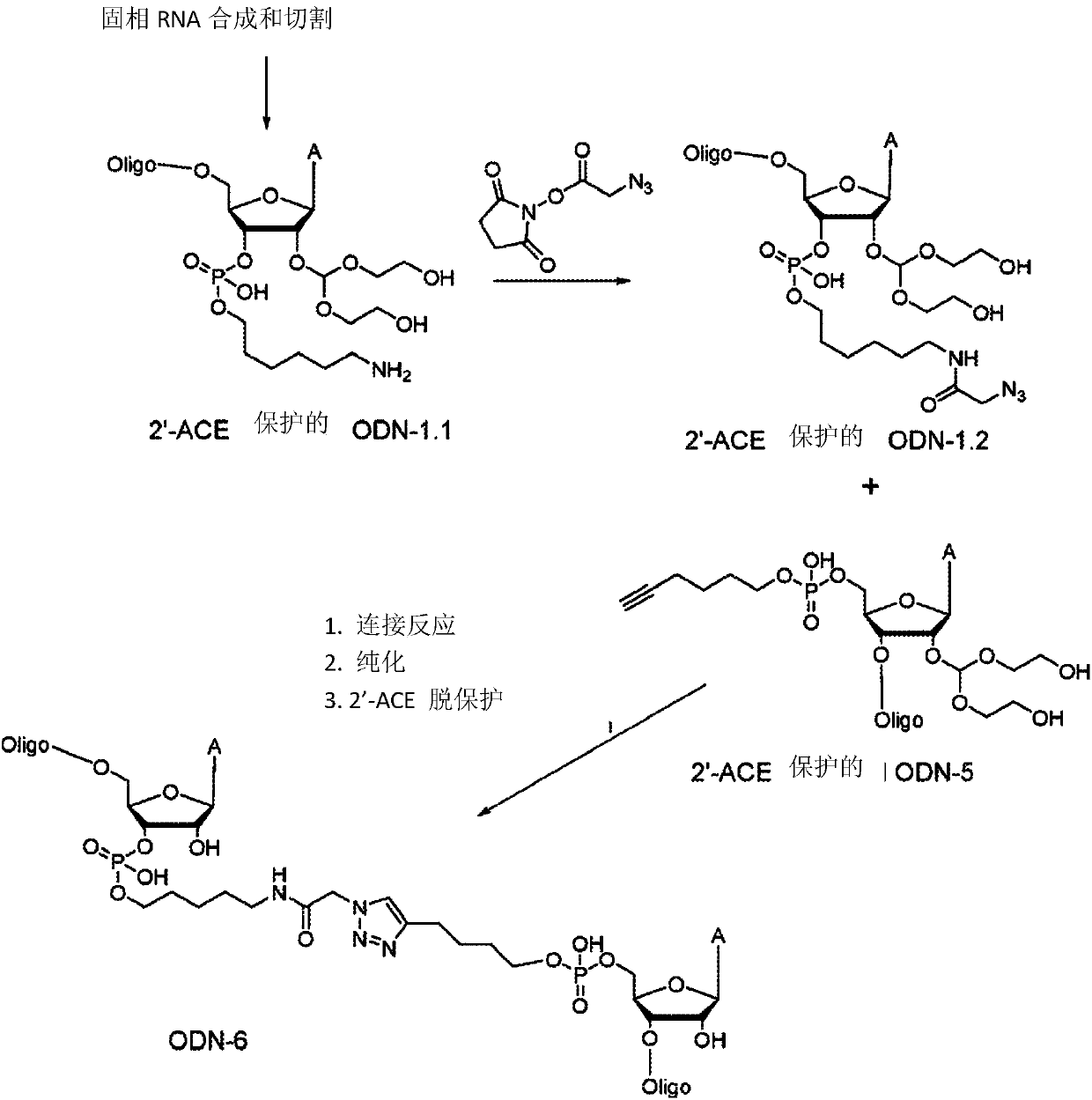
[0093] Example 3. Conjugated oligonucleotide synthesis (Table 1 and image 3 )
[0094] 2'-ACE protected RNA oligonucleotides (ODN-1.1, ODN-2, ODN-3.1, ODN-4, ODN-5, ODN- 7 and ODN-8), using a polystyrene solid support and 2'-bis(acetoxyethoxy)-methyl ether (2'-ACE) phosphoramidite. For ODN-2 and ODN-4, Aminomethylated polystyrene support 6 was used (see Example 1). For ODN-5, 5'-hexyne phosphoramidite 8 was used. After completion of the synthesis cycle, the oligonucleotides on the support were placed on Na at room temperature 2 S 2 solution, followed by washing with water. Oligonucleotides were cleaved from the support with 40% aqueous N-methylamine (NMA), heated at 55°C, and then freeze-dried. Crude RNA was desalted, purified by HPLC, and the identity of the purified samples was confirmed by UPLC and ESI-MS.
[0095] ODN-1.2 and ODN-3.2: After synthesis, NHS azidoacetate (Click ChemicalTools) dissolved in DMF was added to lyophilized 3'-aminoalkyl-modified oligonucleot...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com