Preparation method and application of alginatelyase OalC6
A technology of alginate lyase and construction method, which can be applied in the directions of lyase, carbon-oxygen lyase, and botanical equipment and methods, and can solve problems such as the existence of food demand pressure
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
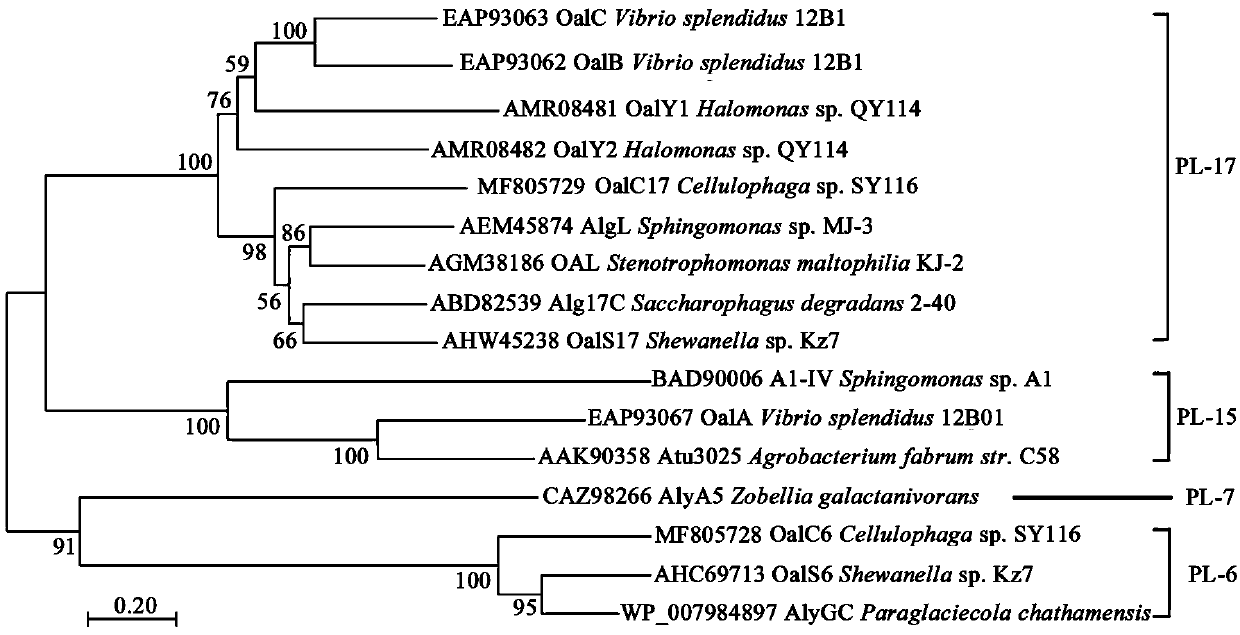
[0026] Example 1: Cloning and identification of the gene encoding oligofucoidan lyase OalC6
[0027] According to Cellulophaga The possible oligofucoidan lyase gene in the sp. SY116 genome was designed with the following amplification primers: upstream primer (5'-CAAAAACATACATTATAC -3') and downstream primer (5'- CTAATTAATCCTAAATTTTT-3'). Using the extracted strain genome as a template, PCR amplification was performed to obtain the full-length sequence of the oligofucoidan lyase OalC6 gene. The PCR conditions were as follows: pre-denaturation at 94 °C for 5 min, followed by 30 cycles of 94 °C for 30 s, 54.8 °C for 30 s, and 72 °C for 1 min 30 s, and finally extended at 72 °C for 10 min.
[0028] After gene sequencing, the full-length 2,328 bp nucleotide sequence of the oligofucoidan lyase OalC6 gene was obtained, and the nucleotide sequence is shown in SEQ ID NO.1; encoding 775 amino acids, and the amino acid sequence is shown in SEQ ID NO.2 As shown, the theoretical molecu...
Embodiment 2
[0030] Example 2: Construction of oligofucoidan lyase OalC6 recombinant expression vector
[0031] The upstream primer (5'-CAAAAACATACATTATAC-3') and the downstream primer (5'-CTAATTAATCCTAAATTTTT-3') were designed according to the complete sequence of the oligofucoidan lyase gene. PCR amplification was carried out to obtain the full-length sequence of alginate lyase OalC6 gene. PCR conditions were: 94°C pre-denaturation for 5 min, followed by 30 cycles of 98°C for 30 s, 58°C for 30 s, 68°C for 2 min, and finally extension at 68°C for 10 min. PCR product and Escherichia coli expression vector pET28a(+) were digested, connected and transformed into E. coli For DH5α competent cells, single clones were picked and cultured in LB liquid containing resistance, plasmids were extracted and digested to identify positive clones. This recombinant plasmid was named pET28a-oalC6. Transform positive plasmids into expression hosts E. coli BL21 competent cells.
Embodiment 3
[0032] Example 3: Recombinant expression and purification of oligofucoidan lyase OalC6 gene in Escherichia coli
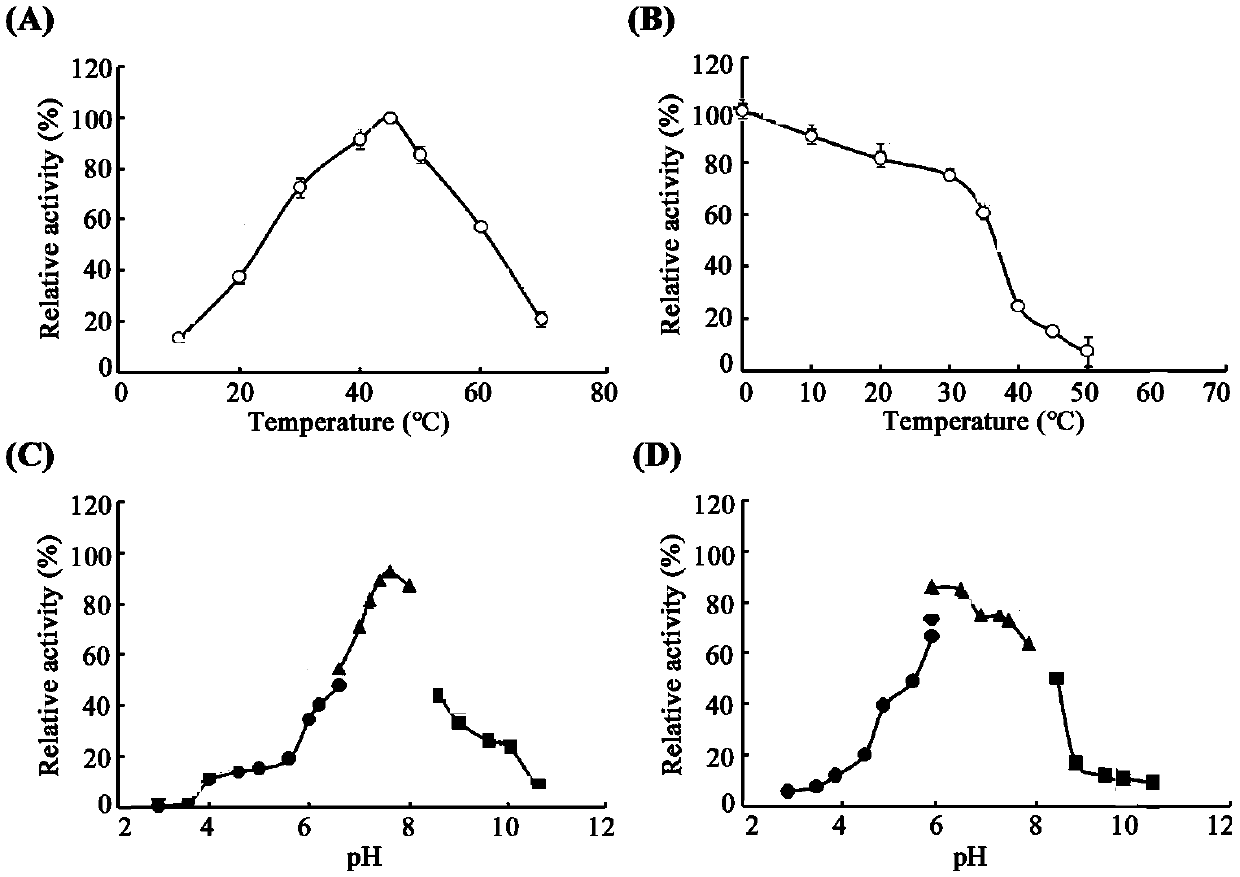
[0033] Transform the recombinant expression plasmid pET28a-oalC6 into the expression host E. coli BL21 competent cells. will reorganize E. coli Pick a single clone of BL21 bacteria into the LB liquid with the same resistance, and culture it with shaking at 37 °C until OD 600 At about 0.6, IPTG was added for induction (final concentration: 0.5 mM), and expression was induced at 25°C for 20 h. The target protein was separated and purified by Ni affinity chromatography. The purification of oligofucoidan lyase OalC6 was detected by polyacrylamide gel electrophoresis, and the results were as follows figure 2 As shown, the purified OalC6 presents a single band on the electrophoresis gel, and the position is consistent with the predicted molecular weight.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com