SNP marker related to enterotoxigenic Escherichia coli F4 type piglet diarrhea resistance and detection method
A technology for Escherichia coli and piglet diarrhea, applied in the field of molecular biology, can solve the problems of inability to determine the causal variation sites of key genes, difficulty in direct application of piglet diarrhea resistance selection, and no reliable conclusion on the regulation mechanism of Escherichia coli F4 resistance. , to achieve the effect of important economic and social value
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] 1. Source of experimental animals
[0030] Collected from the Changping Experimental Pig Farm of the Chinese Academy of Agricultural Sciences.
[0031] There are 79 Landrace pigs, 203 Large White pigs, and 86 Songliao black pigs, including 301 piglets, a total of 348.
[0032] 2. Adhesion phenotype determination
[0033] After 35 days of age, the small intestinal epithelial tissue was slaughtered and collected for the adhesion test of Escherichia coli F4, which were divided into two types: adherent and non-adherent.
[0034] 3. Genomic DNA extraction
[0035] Pig ear tissue samples were collected, placed in centrifuge tubes filled with 70% alcohol, and stored in a -20°C refrigerator for later use.
[0036] Use the animal blood / tissue genomic DNA extraction kit to extract genomic DNA, the required reagents include:
[0037] Lysate laboratory equipment
[0038] Proteinase K (Germany MERCK Biotechnology Co., Ltd.)
[0039] Blood / cell / tissue genomic DNA extraction kit...
Embodiment 2
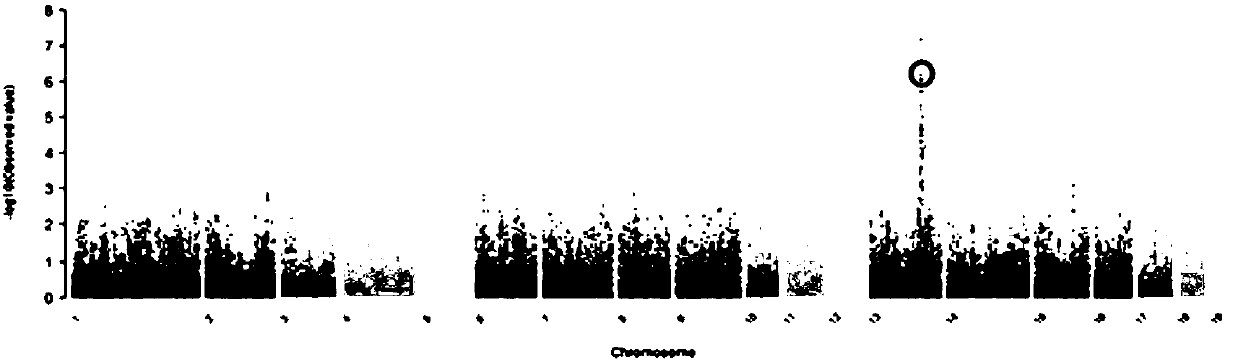
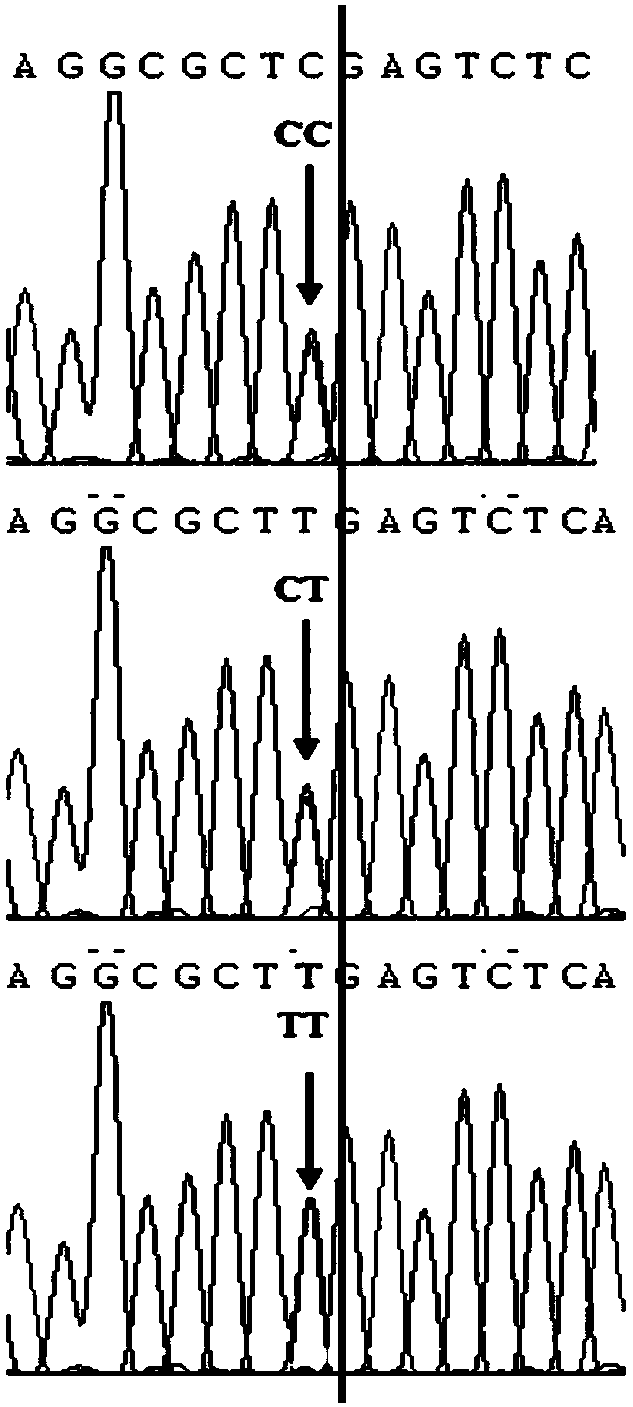
[0062] This example is the verification of the SNP site g.145109683 C / T obtained in Example 1 in Landrace, Large White and Songliao black pig populations.
[0063] 1. Extraction of pig genomic DNA
[0064] The ear tissue samples of three breeds of pigs with adhesion records were collected, placed in centrifuge tubes filled with 70% alcohol, and stored in a -20°C refrigerator for later use. Genomic DNA of pig ear tissue was extracted by the above method, and after quality and concentration detection, the concentration was diluted to 50 ng / μL and stored at -20°C for future use.
[0065] 2. PCR amplification and sequencing of the target fragment
[0066] Use the extracted pig DNA as a template, and perform PCR amplification according to the designed primers: the PCR amplification reaction system is: 1.0 μL of DNA template, 1.0 μL of primers shown in SEQ ID NO: 2 and SEQ ID NO: 3, Taq enzyme reagent 0.125 μL, double distilled water 17.38 μL; The reaction program of PCR amplifica...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com