Primer group and kit
A primer set and kit technology, applied in recombinant DNA technology, microbial determination/inspection, biochemical equipment and methods, etc., can solve the problem of high probe cost, high difficulty of multiplex fluorescent PCR, and inability to achieve 6 STS amplification. Increase the number of problems, to achieve the effect of shortening time-consuming and easy operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0050] Embodiment 1 solution composition, configuration
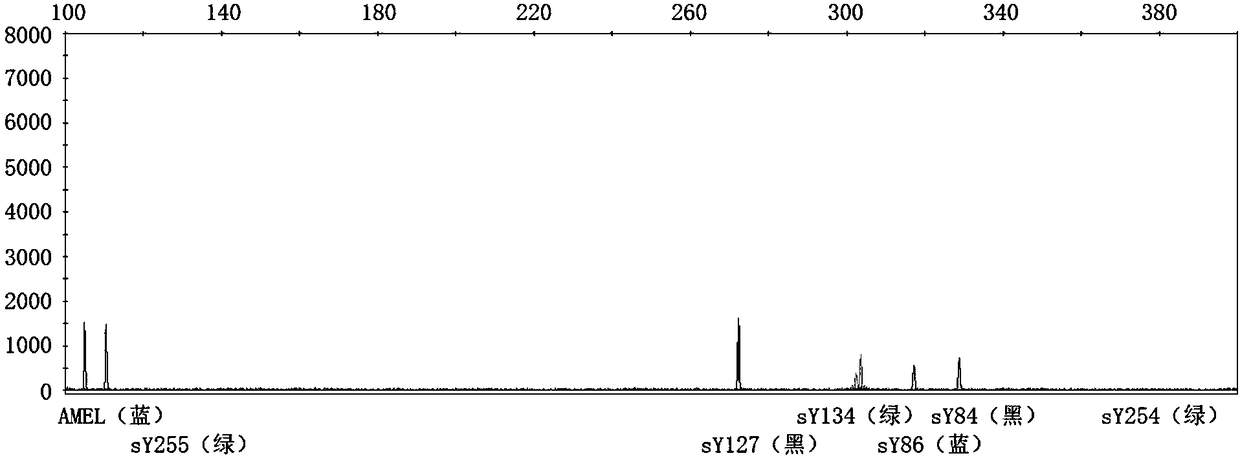
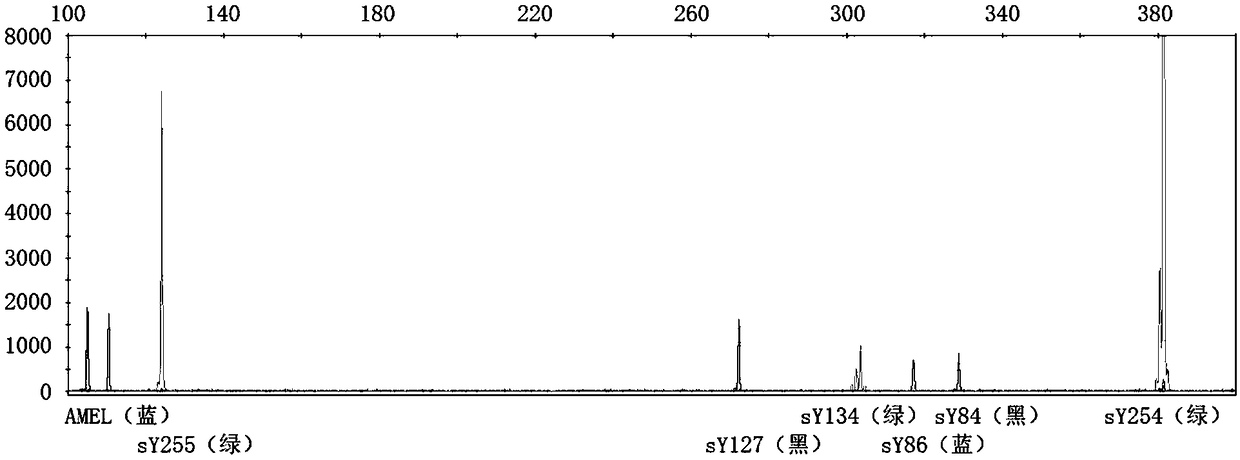
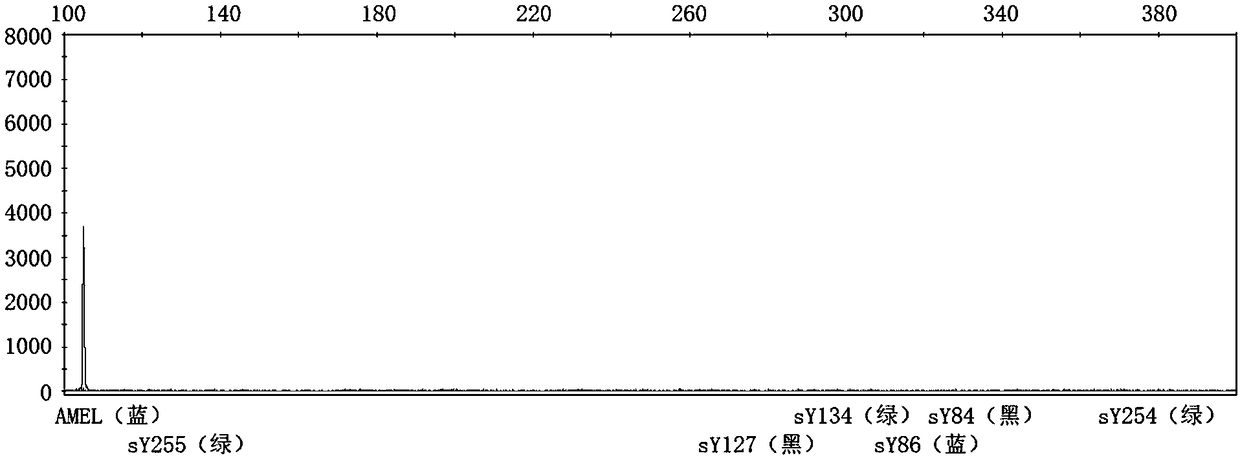
[0051] (1) According to the primer information published in "EAA EMQN best practical guidelines for molecular diagnosis of Ychromosomal microdeletions state of the art 2013", the primers were synthesized by the primer synthesis factory. The forward primer in the primer pair adds a fluorophore at the 5' end. The fluorescent color and primer information are shown in Table 1.
[0052] Table 1: STS primer sequences and fluorescent modifications
[0053]
[0054]
[0055] (2) After obtaining the primer powder synthesized by the primer synthesis manufacturer, centrifuge at 12000rpm for 5min, and use nucleic acid-free ddH 2 O dissolves the primers. According to the primer unit on the primer information sheet, dilute to a primer stock solution with a molar concentration of 100 pmol / μL.
[0056] Vortex and mix well, then centrifuge at low speed to collect the solution at the bottom of the tube.
[0057] (3) Prepare PC...
Embodiment 2
[0063] Embodiment 2PCR operation
[0064] (1) Thaw the PCR reaction solution, enzyme solution, positive quality control, negative quality control, female quality control and blank quality control solution prepared in Example 1, vortex and mix for 5 seconds, and centrifuge at low speed for 10 seconds before use. According to the form of "specimen number + 5" required for the experiment, draw the PCR reaction solution based on 18.8 μL, and put it into a 1.5mL EP tube.
[0065] (2) Pipette 0.2uL of enzyme solution into the above-mentioned EP tube for mixing. Vortex for 5s, centrifuge at low speed for 10s and set aside
[0066](3) Pipette 19 μL of the mixture into a 0.2mL PCR tube, or 0.2mL 8-strip tube. At the same time, add 1 μL of the sample to be tested (be sure to dilute the sample DNA so that the concentration is 5-10ng / μL), and cover the tube cap. Add 1 μL of quality control DNA solution in the same way. Vortex for 5 s and centrifuge at low speed for 10 s.
[0067] (4)...
Embodiment 3ABI3500
[0070] Example 3 ABI3500 computer operation
[0071] (1) Vortex and mix the PCR product of Example 2 for 5 s, centrifuge at low speed for 10 s, and set aside.
[0072] (2) With the ratio of 9.5 μL HiDi fomamide and 0.5 μL Genescan-500 (LIZ) SIZE STD KIT, mix the solution according to the form of “PCR product number + 1”. Vortex for 5 s and centrifuge at low speed for 10 s.
[0073] (3) Pipette 10 μL of the above solution into a new 0.2mL PCR tube or 0.2mL 8-strip tube, add 1 μL of the product to it at the same time, and cover the tube cap. Vortex for 5 s and centrifuge at low speed for 10 s.
[0074] (4) Put the PCR tube into the PCR instrument, set the program: 94°C for 5 minutes, and immediately transfer to ice for cooling for 5 minutes after the end.
[0075] (5) Using the 96-well plate matched with the ABI Genetic Analyzer, transfer the solution described in (4) into the wells of the 96-well plate one by one.
[0076] (6) Cover with the matching plug of ABI Genetic Ana...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com