SNP (single nucleotide polymorphism) molecular marker related to watermelon peel background color and application of SNP molecular marker
A molecular marker and molecular marker-assisted technology, which is applied in application, recombinant DNA technology, microbial measurement/inspection, etc., can solve the problems of few molecular markers, low breeding efficiency, difficulty in obtaining breakthrough new varieties, etc., and achieve accelerated breeding Process, high accuracy, and the effect of improving accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0018] Example 1 Obtaining of SNP sites closely linked to watermelon peel background color traits
[0019] 1.1. BSA of hybrid population (mixed pool resequencing)
[0020] Using a yellow-peeled watermelon (94E1) as the female parent and a green-peeled watermelon (Qingfeng) as the male parent for cross-breeding, an F2 segregating population containing watermelon with yellow rind background and watermelon with green rind background was prepared, and the population was separated from F2 generation. Select 30 watermelons each with a yellow peel background and a green peel background, extract the leaf genomic DNA, and mix the genomic DNA of 30 watermelons with yellow peel background and 30 watermelons with green peel background. The DNA is mixed together. It was sequenced with the Illumina HiSeq 2000 sequencer to obtain 33.75G data, which was compared with the watermelon reference genome 97103 (http: / / cucurbitgenomics.org / organism / 1). 247012 SNP loci were identified, and the SNP-index...
Embodiment 2
[0027] Example 2 Using SNP molecular markers to identify watermelon peel background color
[0028] 2.1, DNA extraction
[0029] Use conventional CTAB method to extract genomic DNA from watermelon leaves and remove RNA. The volume of genomic DNA sample is 50μL. Measure the OD value of genomic DNA sample at 260nm and 280nm with ultraviolet spectrophotometer, and calculate DNA content and OD 260 / 280 Ratio, select OD 260 / 280 Genomic DNA samples with values between 1.8 and 2.0, diluted to 100ng / μL.
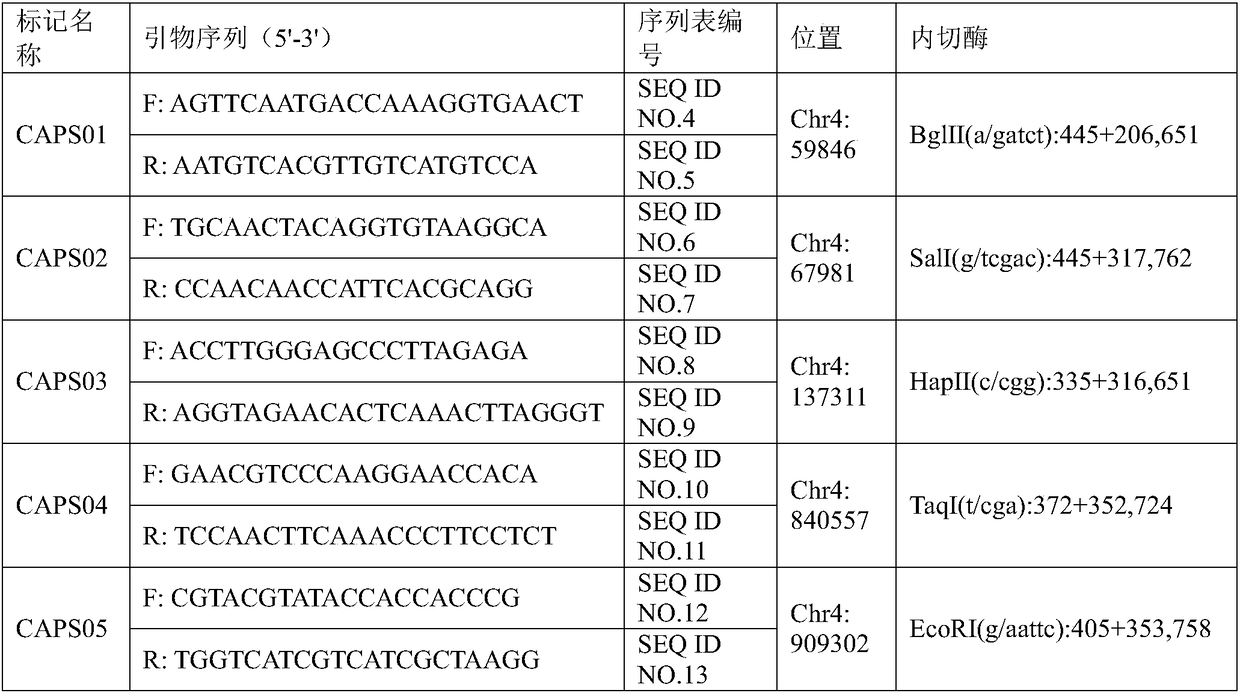
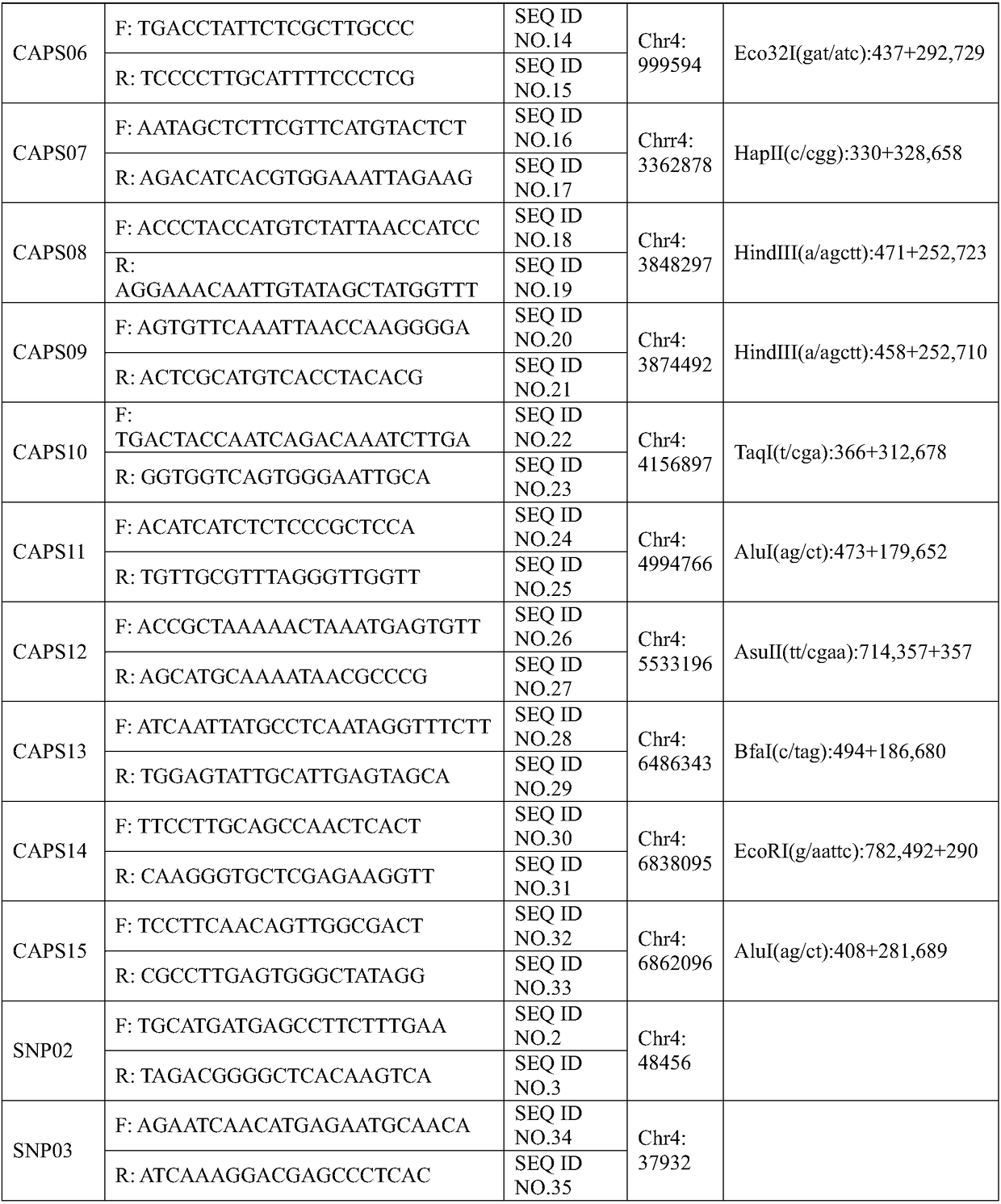
[0030] 2.2, primer
[0031] The primers are the primer pair of SNP molecular marker (SNP02) obtained in Example 1 closely linked to the background color traits of watermelon peel. The primer pair is designed based on the upstream and downstream sequence of the 48456th nucleotide of chromosome 4 of the watermelon genome. The primer sequences are shown in Table 2. The primers were synthesized by Shenggong Bioengineering (Shanghai) Co., Ltd. and diluted to 10μM for use.
[0032] Table 2 Primer ...
Embodiment 3
[0045] Example 3 Using SNP molecular markers to identify the peel background color of the hybrid population
[0046] 3.1. Selection of experimental materials
[0047] The 618 individual plants of the F2 population of the cross combination 94E1 (yellow) × Qingfeng (green) were used as experimental materials, of which 94E1 (yellow) was the female parent and Qingfeng (green) was the male parent. The phenotypic test results showed that the F2 population Among them, there are 458 individual plants with yellow skin background and 160 individual plants with green skin background.
[0048] 3.2. Use SNP molecular markers to identify F2 populations
[0049] The SNP molecular marker (SNP02) related to the peel background color on the watermelon 4 obtained in Example 1 was used to identify 618 F2 individual plants. The specific identification method refers to the method in Example 2. The identification result shows that the identification result of the SNP molecular marker is consistent with the...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com