RT-LAMP detection primer group and kit for identifying H7 subtype HPAIV and LPAIV and application thereof
A technology of RT-LAMP and detection primers, which is applied in the determination/testing of microorganisms, DNA/RNA fragments, microorganisms, etc. It can solve the problems of long time consumption and unsuitable for grass-roots application, and achieve the effect of simple operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0022] Example 1 :Design and synthesis of primers
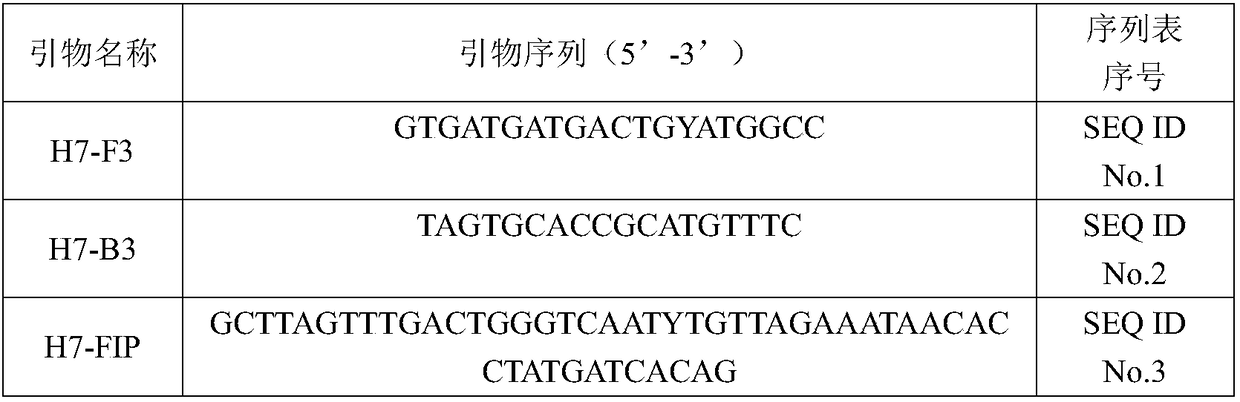
[0023] Download the HA gene sequence of H7 subtype AIV in GeneBank, use MEGA6.0 software for sequence comparison, and use the online software Primer Explorer V4 to design two sets of LAMP primers respectively according to the conservative and specific sequences of H7 subtype AIV. For the detection of H7 subtype and H7 subtype LPAIV, the primer sequences are shown in Table 1.
[0024] Table 1 H7 subtype AIV and H7 subtype LPAIV detection primer sequence
[0025]
[0026]
Embodiment 2
[0027] Example 2: Establishment and optimization of RT-LAMP detection system
[0028] 2.1 Viral nucleic acid extraction and cDNA synthesis
[0029] Refer to the manual of RNA extraction reagent RNAIso plus to extract viral allantoic fluid RNA, and finally dissolve RNA with DEPC water for reverse transcription. 50μL reverse transcription system: 5× buffer 10μL, dNTP (10mmol / L) 2μL, reverse transcription primer 1.5μL (concentration), AMV reverse transcriptase 1μL (10U / μL), RNA inhibitor 0.5μL (40U / μL), 35 μL of viral RNA. After mixing, act at 42°C for 30 minutes.
[0030] 2.2 Preparation of standards
[0031] Use the two pairs of primers H7-F3, H7-B3, H7-F3, H7-B3 in Table 1 to amplify the target fragments of the H7 subtype AIV HA and NA genes, run the PCR product and cut the target fragments after electrophoresis, and connect the vector pEASY, transform DH5α E. coli competent cells, use Amp-containing LB plates to screen E. coli, pick positive colonies for PCR identification, and ...
Embodiment 3
[0035] Example 3: RT-LAMP specificity and sensitivity test
[0036] Using optimized reaction conditions, different subtypes of AIV samples were tested to test the specificity of the RT-LAMP method. The prepared standard was amplified by the established method to test its sensitivity.
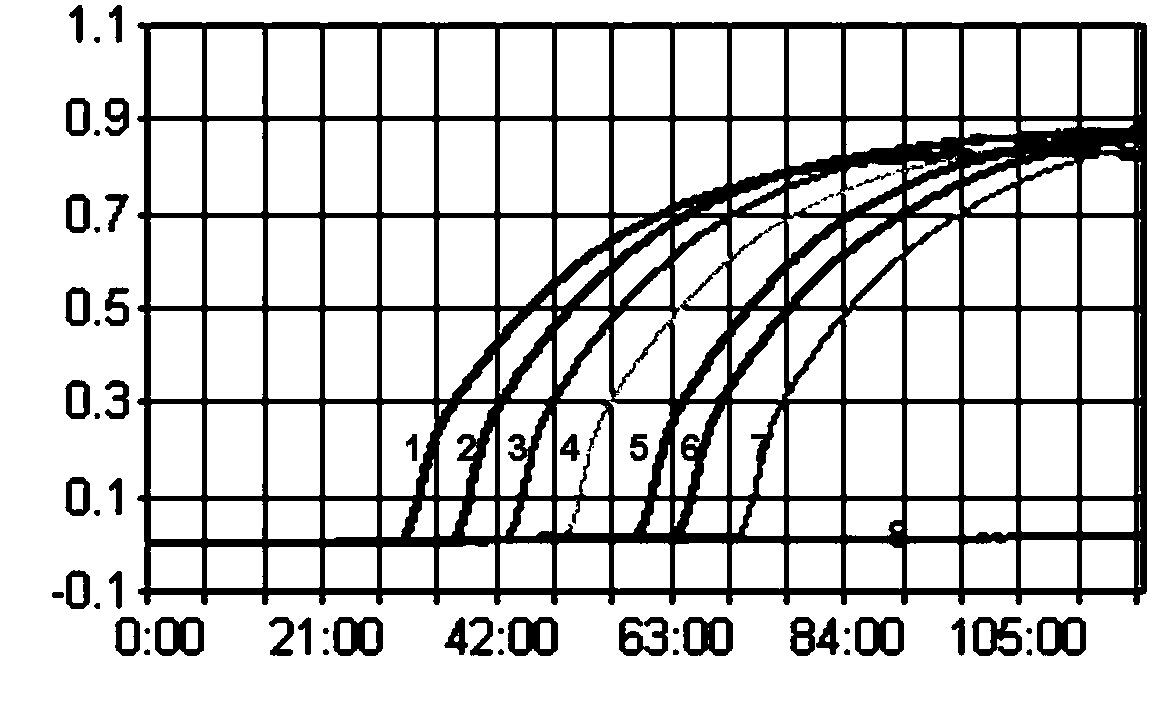
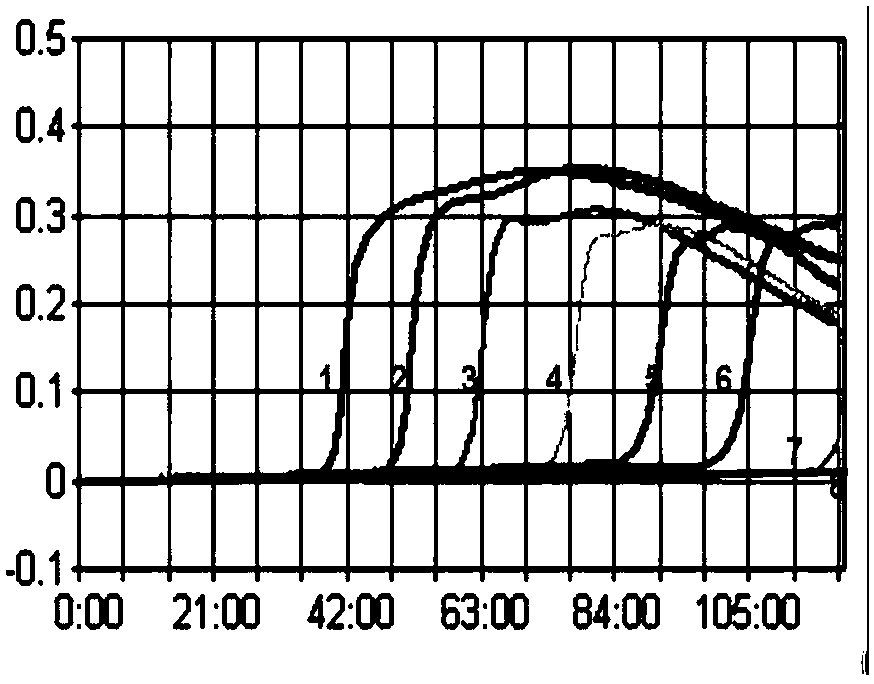
[0037] Such as Figure 1-2 As shown, the established H7 subtype AIV LAMP can detect a standard of 10 copies / μL; the established H7 subtype LPAIV LAMP can detect a standard of 10 copies / μL.
[0038] Table 2 RT-LAMP specific detection
[0039]
[0040] As shown in Table 2, the RT-LAMP test results of H7 subtype AIV RNA were positive, and the test results of other HA subtypes AIV were negative.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com