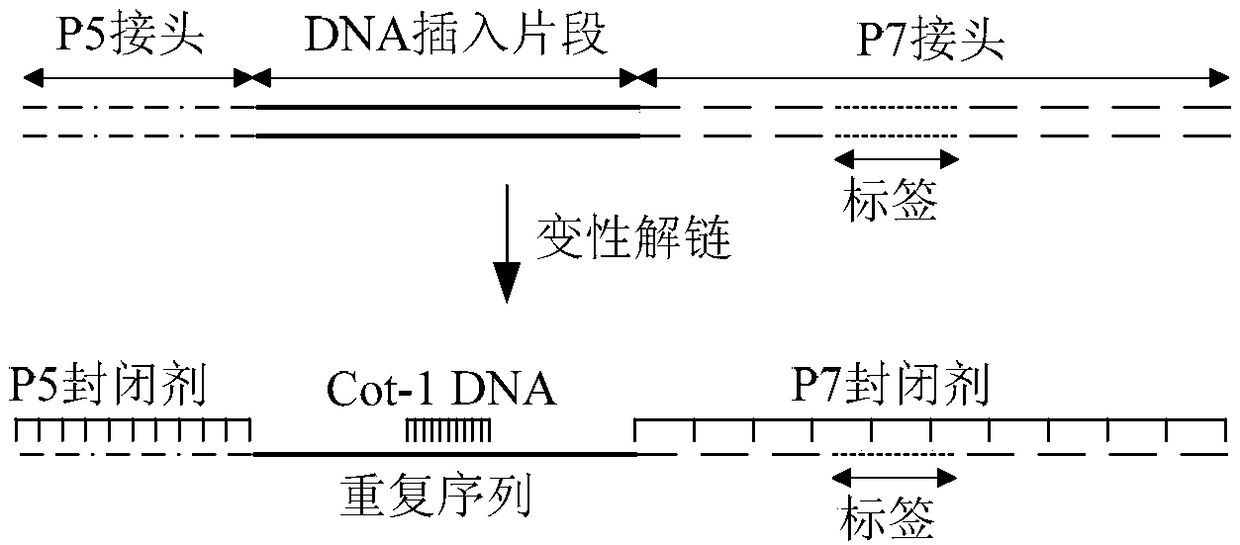
Joint enclosing sequence, library construction kit, and construction method of sequencing library
A technology of adapter blocking sequences and blocking reagents, which is applied in the field of sequencing library construction, can solve the problem of low capture efficiency of target fragments, achieve the effects of reducing non-target region capture, increasing binding efficiency, and improving capture efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
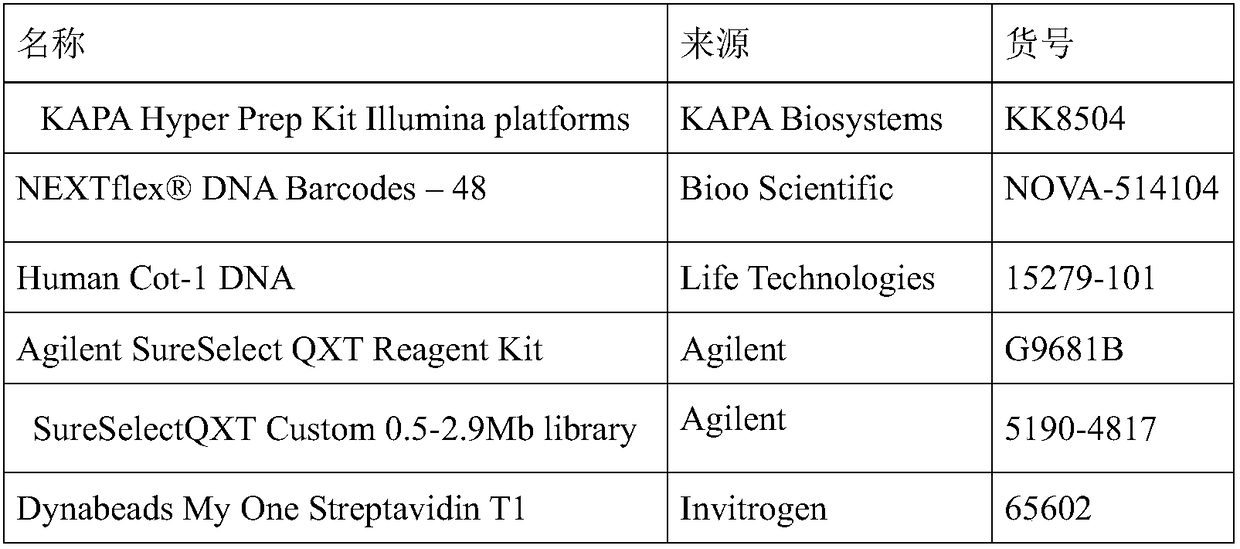
[0041] The reagents used in Example 1 are shown in Table 1, wherein the hybrid capture reagent is the RNA hybrid capture system of Agilent.
[0042] Table 1:
[0043]
[0044] 1. Sample extraction
[0045] Tissue samples were extracted using a paraffin-embedded tissue DNA extraction kit (article number Dp331) from Tiangen Company, and the extracted DNA was stored at -20°C for later use. See Table 2 for sample names and extraction volumes.
[0046] Table 2:
[0047]
[0048] 2. Fragmentation of DNA.
[0049] 2.1---DNA interruption
[0050] Take 6 copies of the sample (520ng / part) and use a Covaris disruption instrument (instrument model S220), and perform DNA fragmentation according to the parameters set in Table 3 below. The main peak of the fragmented product was at 200-350bp, and 20ng was taken for electrophoresis detection.
[0051] table 3:
[0052] Maximum incident power
work factor
Energy Transfer Points
processing time
450W
30...
Embodiment 2
[0202] The specific reagents used in Example 2 are shown in Table 18, wherein the hybridization capture reagent is the DNA hybridization capture system of IDT Company.
[0203] Table 18:
[0204]
[0205] 1. Sample extraction
[0206] Cell line samples were extracted using Tiangen’s Magnetic Bead Method Blood Genomic DNA Extraction Kit (Product No. DP329-02), and the extracted DNA was stored at -20°C for later use. See Table 19 for sample names and extraction volumes.
[0207] Table 19:
[0208]
[0209] 2. Fragmentation of DNA.
[0210] 2.1---DNA interruption
[0211] Take 6 copies of the sample (120ng / part) and use a Covaris disruption instrument (instrument model S220), and perform DNA fragmentation according to the parameters set in Table 20 below. The main peak of the fragmented product was at 200-350bp, and 20ng was taken for electrophoresis detection.
[0212] Table 20:
[0213] Maximum incident power
work factor
Energy Transfer Points
...
Embodiment 1
[0369] The result detection of embodiment one and embodiment two:
[0370] Use bwa software to compare the sequencing results of Example 1 and Example 2 with the reference genome. The specific quality control information is shown in Table 35 below, and the capture efficiency of libraries using different blocking reagents is shown in Table 36 below.
[0371] Table 35:
[0372]
[0373]
[0374] Table 36:
[0375]
[0376]
[0377] Attachment: Probe capture ratio = total bases covering the target region / total bases covering the whole genome * 100%.
[0378] From the above description, it can be seen that the above-mentioned embodiments of the present invention have achieved the following technical effects: compared with the unmodified blocking sequence, the present invention adds modification (especially reverse dT modification) at the 3' end The adapter blocking sequence improves the binding efficiency with the adapter, improves the blocking efficiency, reduces th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com