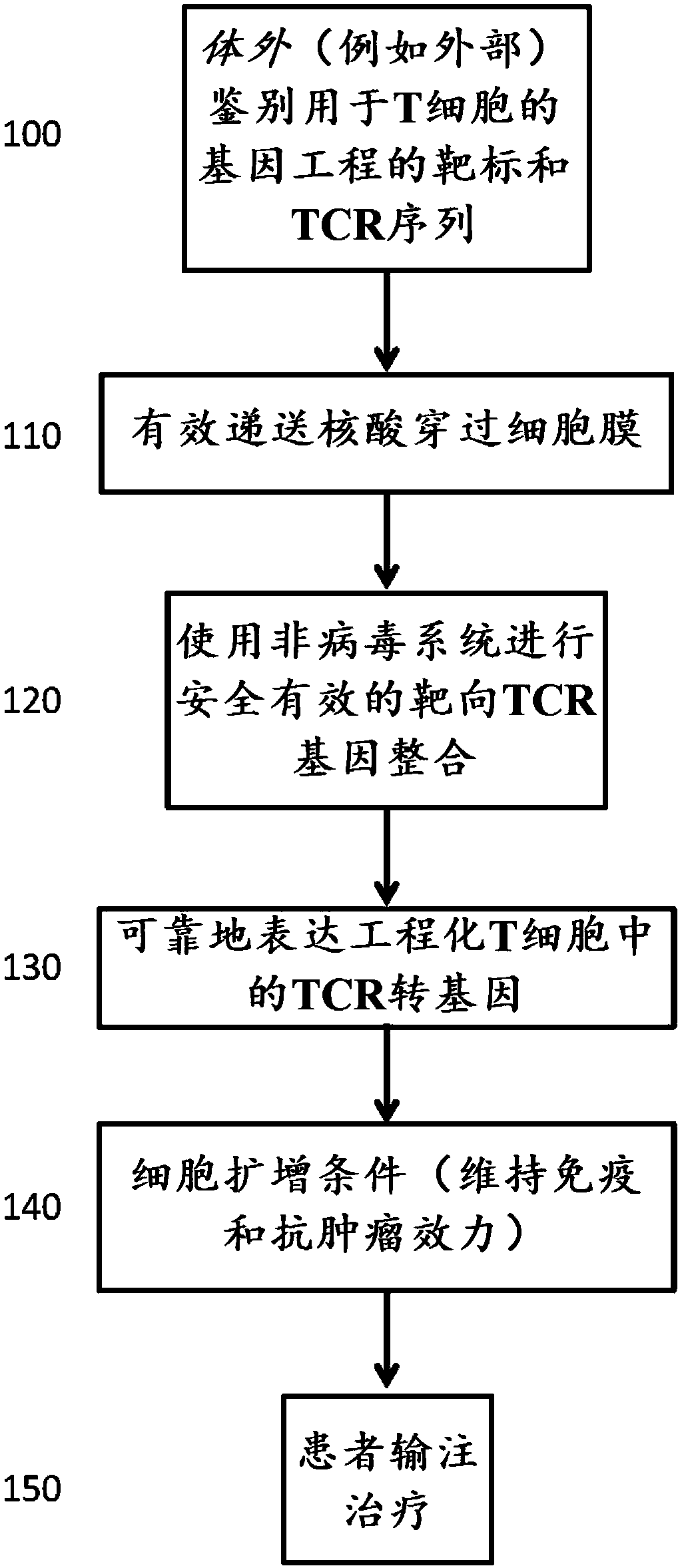
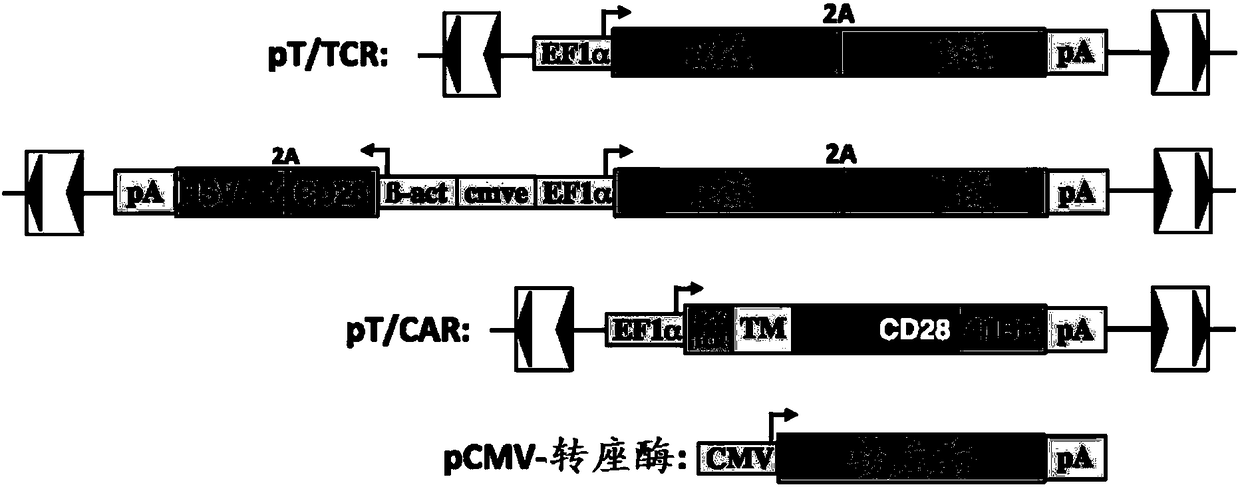
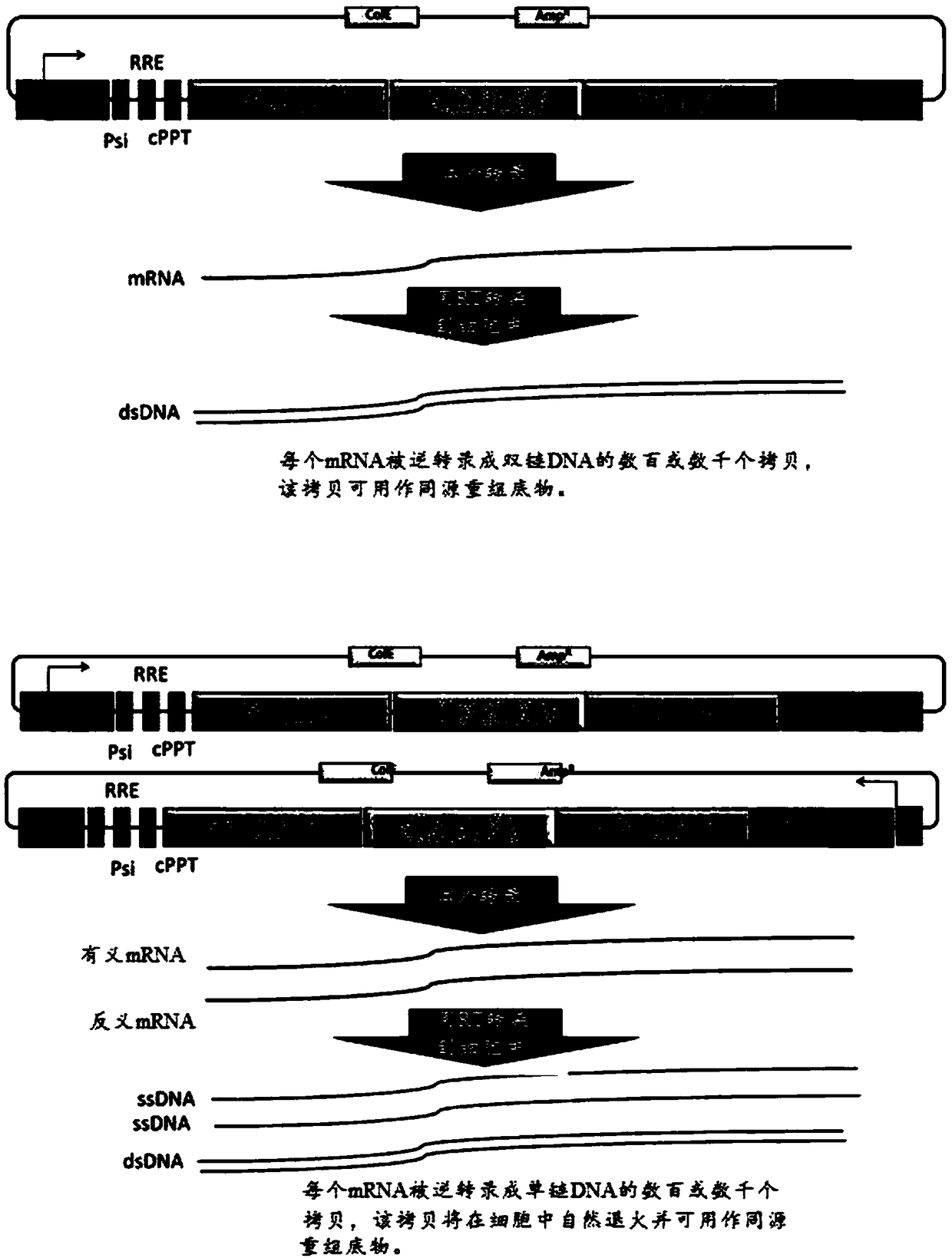
Modified cells and methods of therapy
A technology of cells and lymphocytes, applied in the fields of V-domain immunoglobulins, sequential cell death factor 1, and hepatitis A virus cell receptor 2, which can solve the problems of lack of identifiable molecules and lack of specific binding to tumor targets
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0509] Example 1: Determining the transfection efficiency of various nucleic acid delivery platforms
[0510] Isolation of Peripheral Blood Mononuclear Cells (PBMCs) from LeukoPak
[0511]This article uses leukopak collected from normal peripheral blood. Dilute blood cells 3 to 1 with chilled 1X PBS. Add the diluted blood dropwise (eg, very slowly) onto 15 mL of LYMPHOPREP (stem cell Technologies) in a 50 mL Erlenmeyer flask. Cells were spun for 25 minutes at 400x G with no brake. Slowly remove the buffy coat and place it in a sterile Erlenmeyer flask. Wash the cells with chilled 1X PBS and spin the cells at 400x G for 10 minutes. The supernatant was removed, cells were resuspended in culture medium, counted and viable frozen in freezing medium (45 mL heat-inactivated FBS and 5 mL DMSO).
[0512] Isolation of CD3+ T cells
[0513] PBMCs were thawed and plated in medium (RPMI-1640 (without phenol red), 20% FBS (heat inactivated) and 1X Gluta-MAX) for 1-2 hours. Collect c...
Embodiment 2
[0531] Embodiment 2: Determine the transfection efficiency of GFP plasmid in T cells
[0532] Transfection efficiency of primary T cells nucleoffected with Amaxa using GFP plasmid. Figure 4 The structures of four plasmids prepared for this experiment are shown: Cas9 nuclease plasmid, HPRT gRNA plasmid (CRISPR gRNA targeting human HPRT gene), Amaxa EGFPmax plasmid, and HPRT targeting vector. The HPRT targeting vector has a 0.5kb targeting arm ( Figure 5 ). Sample preparation, flow cytometry, and other methods were similar to Experiment 1. Plasmids were prepared using an endotoxin-free kit (Qiagen). Different conditions (shown in Table 3), including cell numbers and plasmid combinations, were tested.
[0533] Table 3. Different conditions used in the experiments
[0534] Sample ID
#PBMC
plasmid
GFP'(ug)
Cas9'(ug)
gRNA'(ug)
target' (ug)
1
5×10^6
GFP
5
0
0
0
2
2×10^7
Cas9
0.1
20
0
0
3 ...
Embodiment 3
[0537] Example 3: Identification of gRNAs with the highest double-strand break (DSB) induction at each locus
[0538] Design and construction of guide RNA:
[0539] Guide RNAs (gRNAs) were designed to desired gene regions using the CRISPR design program (Zhang Lab, MIT 2015). Various primers (shown in Table 4) for generating gRNAs were selected based on the highest ranking value determined by the off-target position. The gRNAs were sorted by oligonucleotide pairs: 5'-CACCG-gRNA sequence-3' and 5'-AAAC-reverse complementary gRNA sequence-C-3' (sequences of oligonucleotide pairs are listed in Table 4 ).
[0540] Table 4. Primers used to generate gRNA (CACCG sequence added to sense strand and AAAC to antisense strand for cloning purposes).
[0541]
[0542]
[0543]
[0544]The gRNAs were cloned together using the Target Sequence Cloning Protocol (Zhang Lab, MIT). Briefly, using T4PNK (NEB) and 10X T4 Ligation Buffer (T4Ligation Buffer, NEB), oligonucleotide pairs we...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com