Method for screening miR-3880 target gene
A technology of mir-3880 and screening methods, applied in the direction of biochemical equipment and methods, other methods of inserting foreign genetic materials, material inspection products, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
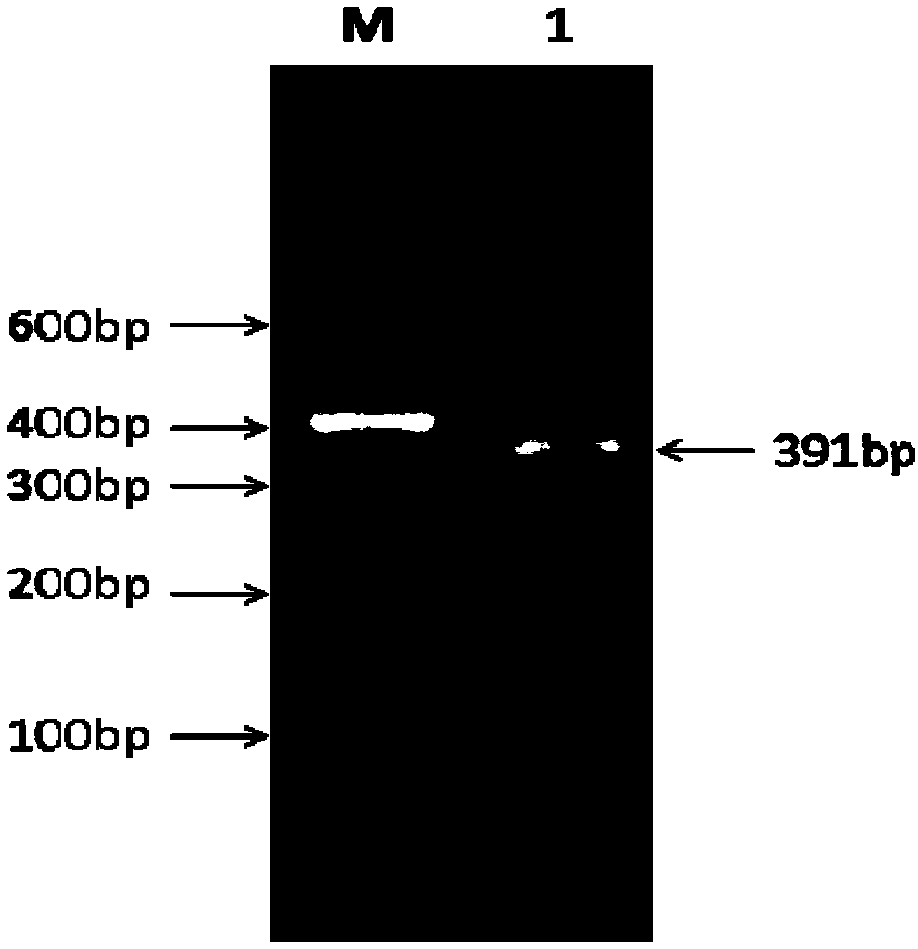
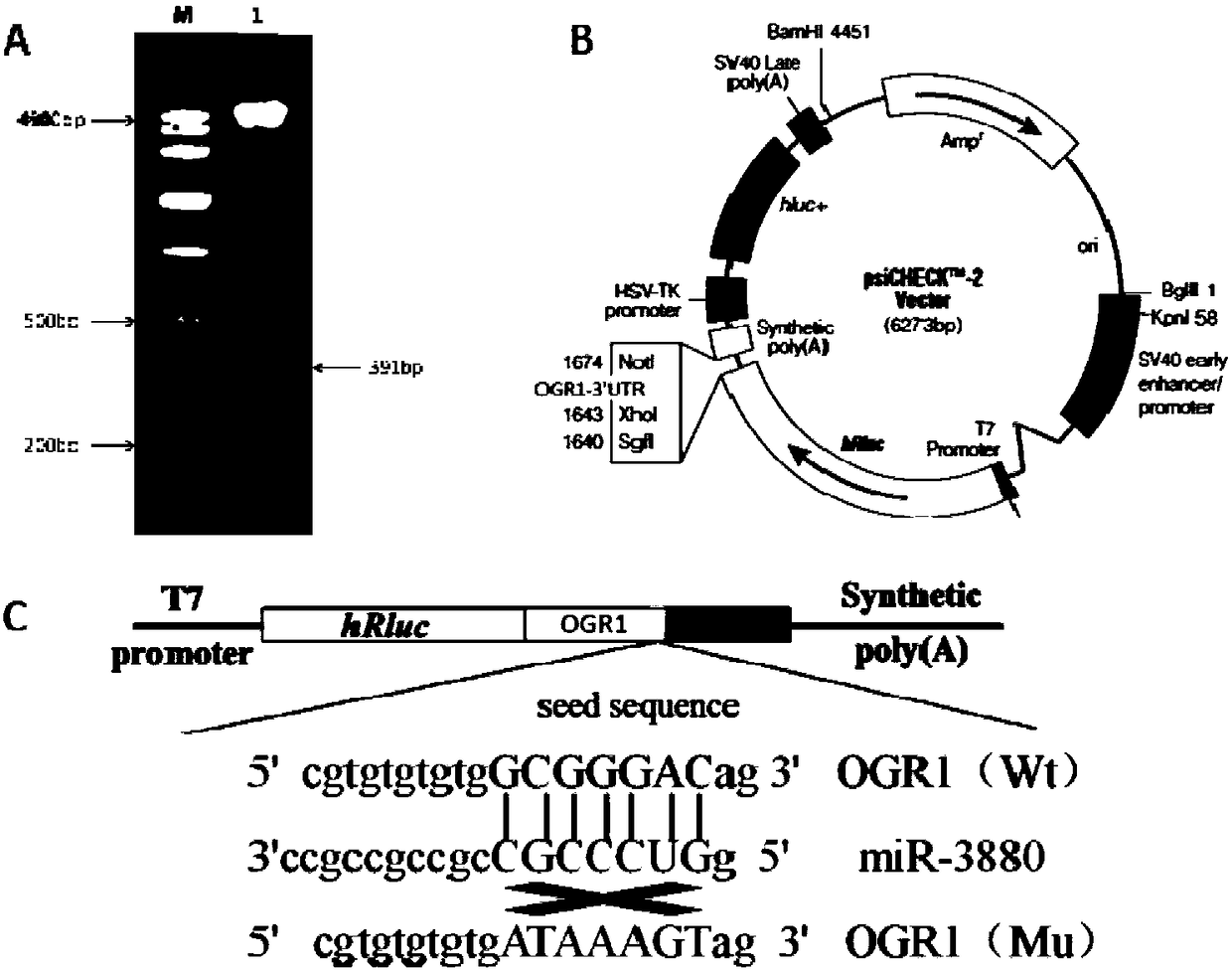
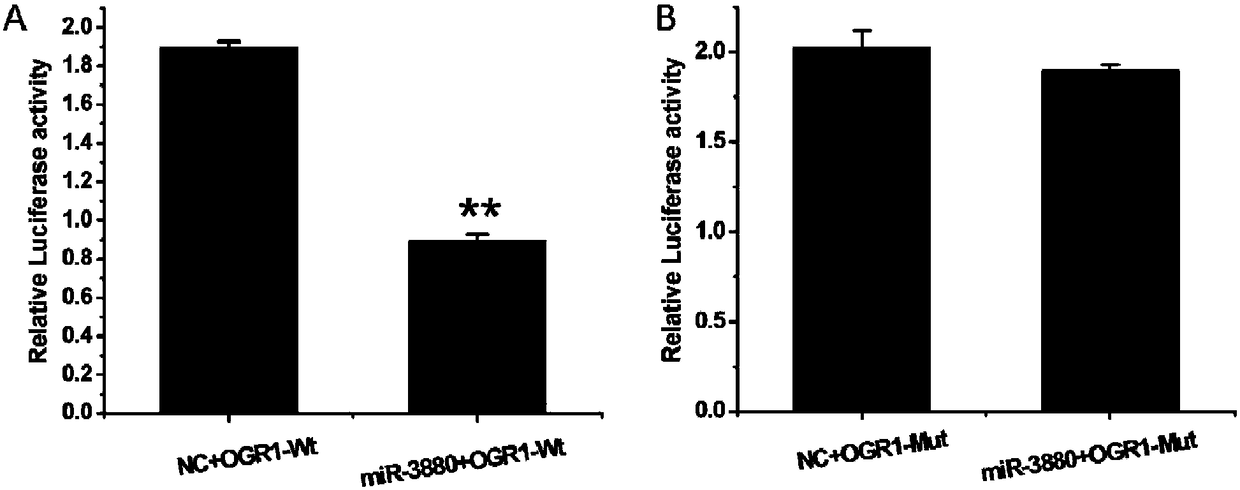
[0067] This embodiment provides a miR-3880 target gene screening method, comprising the following steps:
[0068] 1) Using goat genomic DNA as a template, in Taq DNA polymerase, buffer environment, Mg 2+ 1. In the presence of dNTPs, use primer OGR1 to amplify under PCR conditions, and determine that the obtained PCR product is the sequence of the 3'UTR region of the OGR1 gene;
[0069] The primer OGR1 is as follows (the underline is the restriction site of XhoI and NotI endonuclease):
[0070] Upstream primer F: 5'-CG CTCGA GGTGGAGGTTGGATGGGGAGA-3';
[0071] Downstream primer R: 5'-AT GCGGCCGC ACACATGCACACACAGAGCC-3'.
[0072] The condition of described PCR amplification is:
[0073] The 15 μl reaction system includes: 0.5 μl DNA template, 7.5 μl MasterMix, 0.5 μl each of upstream primer F and downstream primer R of primer OGR1, add sterilized water to 15 μl;
[0074] Described PCR amplification reaction procedure is as follows:
[0075] The PCR reaction program using ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com