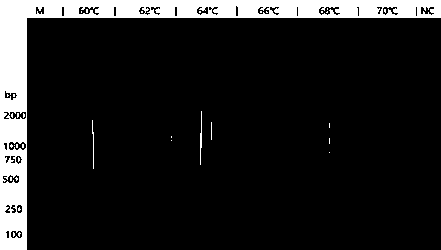LAMP primer group for detecting helicobacter pylori cytotoxin related protein cagA and application thereof
A Helicobacter pylori and cytotoxin technology, applied in the field of biology, can solve the problems of long detection time, insufficient sensitivity, and no symptoms, and achieve the effect of simple operation, high sensitivity, and strong specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
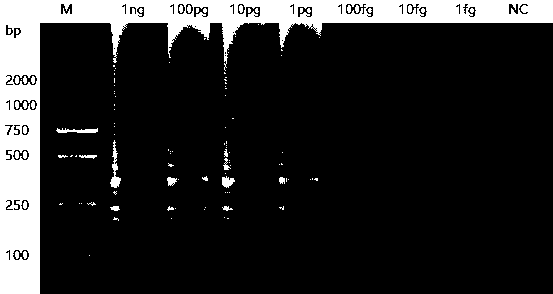
[0033] Cytotoxin-associated protein in Helicobacter pylori ( cagA ) detection limit test
[0034] 1 ng / μL, 100 pg / μL, 10 pg / μL, 1 pg / μL, 100 fg / μL, 10 fg / μL, and 1 fg / μL of HP strain ATCC 700392 DNA were used as templates, respectively.
[0035] LAMP reaction system includes: 1×ThermoPol reaction buffer; 6 mM MgSO4; 0.8 M betaine; 1.4mM dNTPs; the above three pairs of primers, 0.2 μM F3 / B3; 1.6 μM FIP / BIP; 0.8 μM LF / LB; Bst DNA polymerase 320 U / mL; DNA template 1 μL, make up to 20 μL with enzyme-free water. For the blank control group, no DNA was added, and 1 μL of enzyme-free water was added accordingly.
[0036] React in a water bath at 64°C for 30 min, then at 95°C for 5 min to inactivate the enzyme. The reaction product was electrophoresed on a 2% agarose gel. If a ladder-shaped band appeared, it was indicated that there was a LAMP reaction, and if there was no ladder-shaped band, it was indicated as no reaction.
[0037] Detection limit test results: HP strains have...
Embodiment 2
[0039] Cytotoxin-associated protein in Helicobacter pylori ( cagA ) Reaction temperature optimization
[0040] 1 ng of HP strain ATCC 700392 DNA was used as template.
[0041] LAMP reaction system includes: 1×ThermoPol reaction buffer; 6 mM MgSO4; 0.8 M betaine; 1.4 mMdNTPs; the above three pairs of primers, 0.2 μM FIP / BIP; 1.6 μM F3 / B3; 0.8 μM LF / LB; Bst DNA polymerase 320U / mL; DNA template 1 μL, make up to 20 μL with enzyme-free water. For the blank control group, no DNA was added, and 1 μL of enzyme-free water was added accordingly.
[0042] React in water baths at 60°C, 62°C, 64°C, 66°C, 68°C, and 70°C for 30 minutes respectively, and then react at 95°C for 5 minutes to inactivate the enzyme. The reaction product was electrophoresed on a 2% agarose gel. If a ladder-shaped band appeared, it was indicated that there was a LAMP reaction, and if there was no ladder-shaped band, it was indicated as no reaction.
[0043]Reaction temperature optimization results: HP strain ...
Embodiment 3
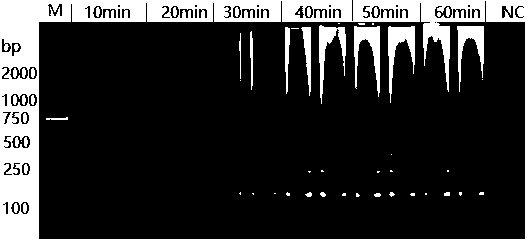
[0045] Optimization of reaction time of cytotoxin-associated protein (cagA) in Helicobacter pylori
[0046] 1 ng of HP strain ATCC 700392 DNA was used as a template.
[0047] LAMP reaction system includes: 1× ThermoPol reaction buffer; 6 mM MgSO 4 ; 0.8 M betaine; 1.4 mMdNTPs; the above three pairs of primers, 0.2 μM F3 / B3; 1.6 μM FIP / BIP; 0.8 μM LF / LB; Bst DNA polymerase 320U / mL; DNA template 1 μL, make up to 20 μL with enzyme-free water. For the blank control group, no DNA was added, and 1 μL of enzyme-free water was added accordingly.
[0048] React in a water bath at 63°C for 10 min, 20 min, 30 min, 40 min, 50 min, and 60 min, respectively, and then react at 95°C for 5 min to inactivate the enzyme. The reaction product was electrophoresed on a 2% agarose gel. If a ladder-shaped band appeared, it was indicated that there was a LAMP reaction, and if there was no ladder-shaped band, it was indicated as no reaction.
[0049] Reaction time optimization results: the produc...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com