Method for preparing chimerism-free gene knock-out animals based on CRISPR/Cas9 technology
A gene knockout and animal technology, applied in the field of molecular biology, can solve the problem of low efficiency of biallelic gene knockout
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
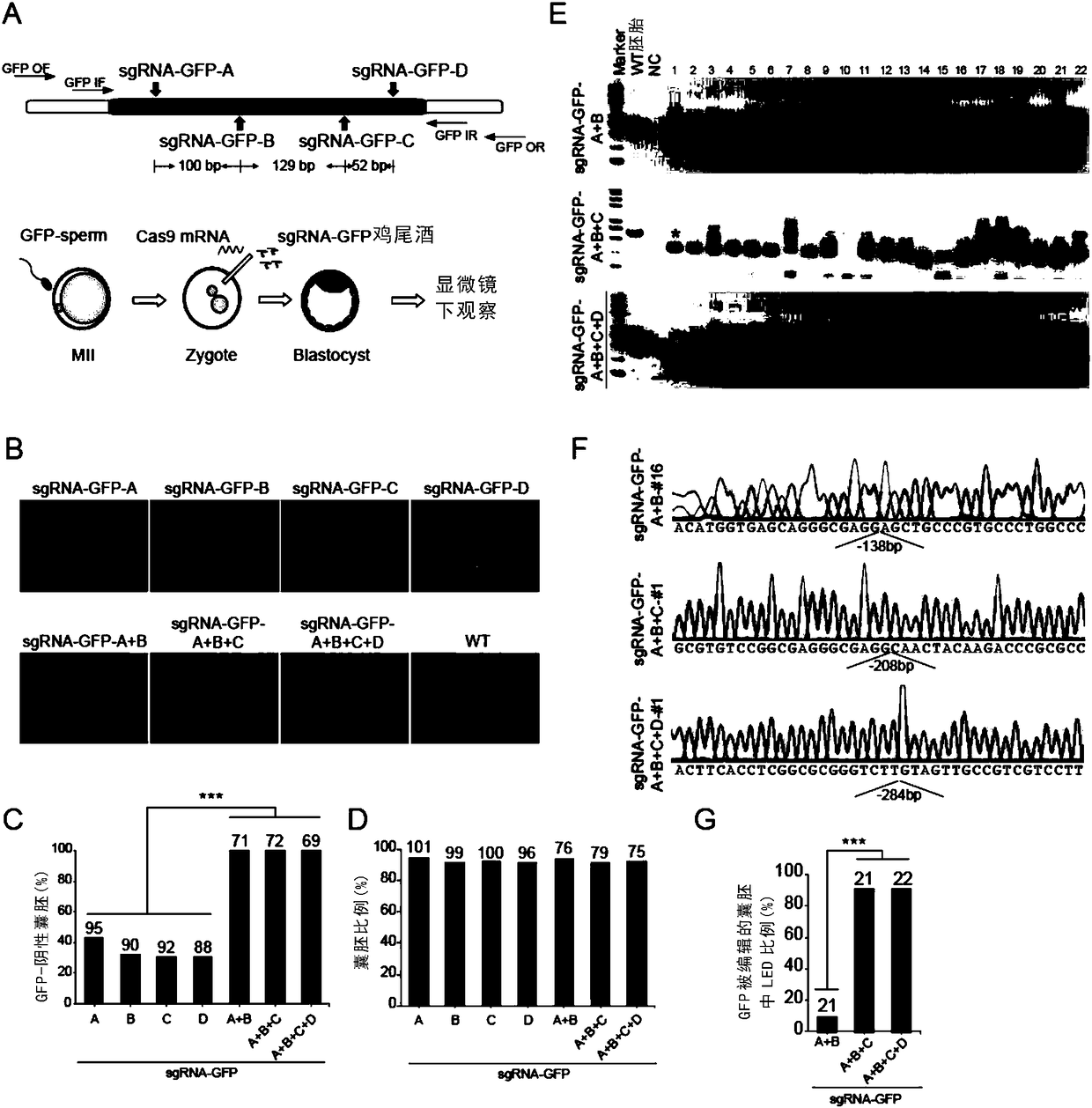
[0219] Example 1. Complete deletion of the green fluorescent gene in the transgenic green fluorescent embryo
[0220] The inventors first wanted to know whether the pre-injection of Cas9 mRNA and sgRNA into mouse MII eggs in fertilized eggs could reduce their chimerism. The targeted genes are Tet1 and Tet2; the primers used to make in vivo transcription templates are shown in the corresponding sequences in Table 1 (Tet1 and Tet2 and sgRNA-R); the sgRNA target sequences are shown in the corresponding sequences in Table 2; the primers used in genotype analysis Such as the corresponding sequence in Table 3. Two-step injection: inject Cas9 mRNA and sgRNA into mouse MII oocytes, and inject sperm into MII oocytes 4 hours later. One-step injection: Inject Cas9 mRNA and sgRNA into mouse zygotic embryos. As a result, no obvious effect was found, and chimerism was not significantly reduced, such as Figure 7 A-F.
[0221] Although the inventor adjusted and designed a variety of sgRN...
Embodiment 2
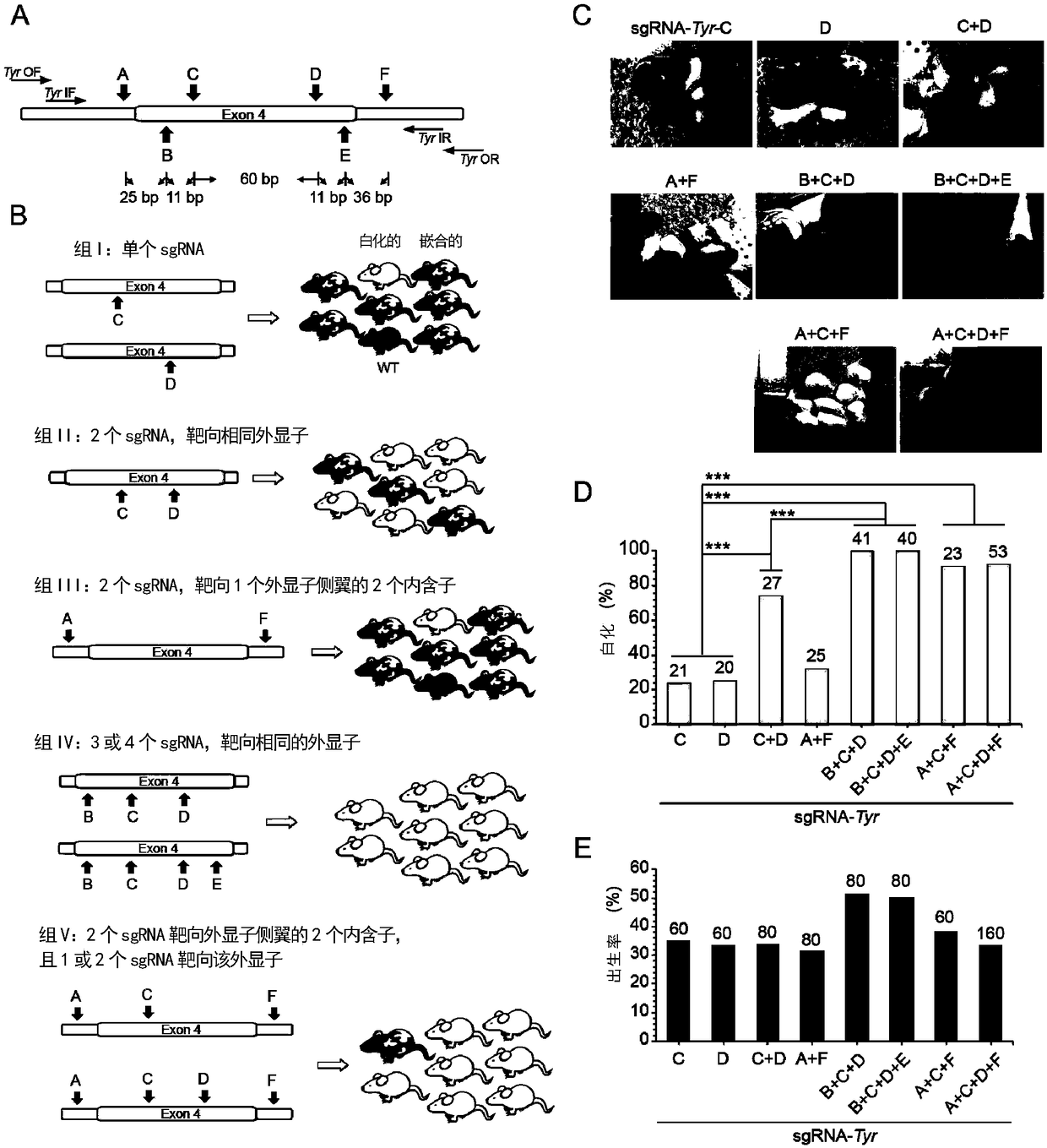
[0225] Embodiment 2, completely delete the Tyr gene of mouse
[0226] In order to further test whether C-CRISPR can completely delete endogenous biallelic genes, the inventors targeted the Tyrosinase gene (Tyr, for pigmentation) of fertilized eggs, and obtained gene editing by two-cell embryo transfer mice ( figure 2 A). Mice with one or two copies of wild-type Tyr appear fully pigmented, whereas mice with both alleles nullified are albino, so chimerism can be seen visually. The primers used to make templates for in vivo transcription are shown in Table 1 for the corresponding sequences (Tyr-A to Tyr-F and sgRNA-R); the sgRNA target sequences are shown in Table 2 for the corresponding sequences. The sgRNA target sites A (Tyr-A), F (Tyr-F) are located in the intron position next to the exon at the two ends of exon 4; the sgRNA target sites B (Tyr-B), C (Tyr- C), D(Tyr-D), E(Tyr-E) are located in exon 4.
[0227] The inventors divided the gene editing experiment into five g...
Embodiment 3
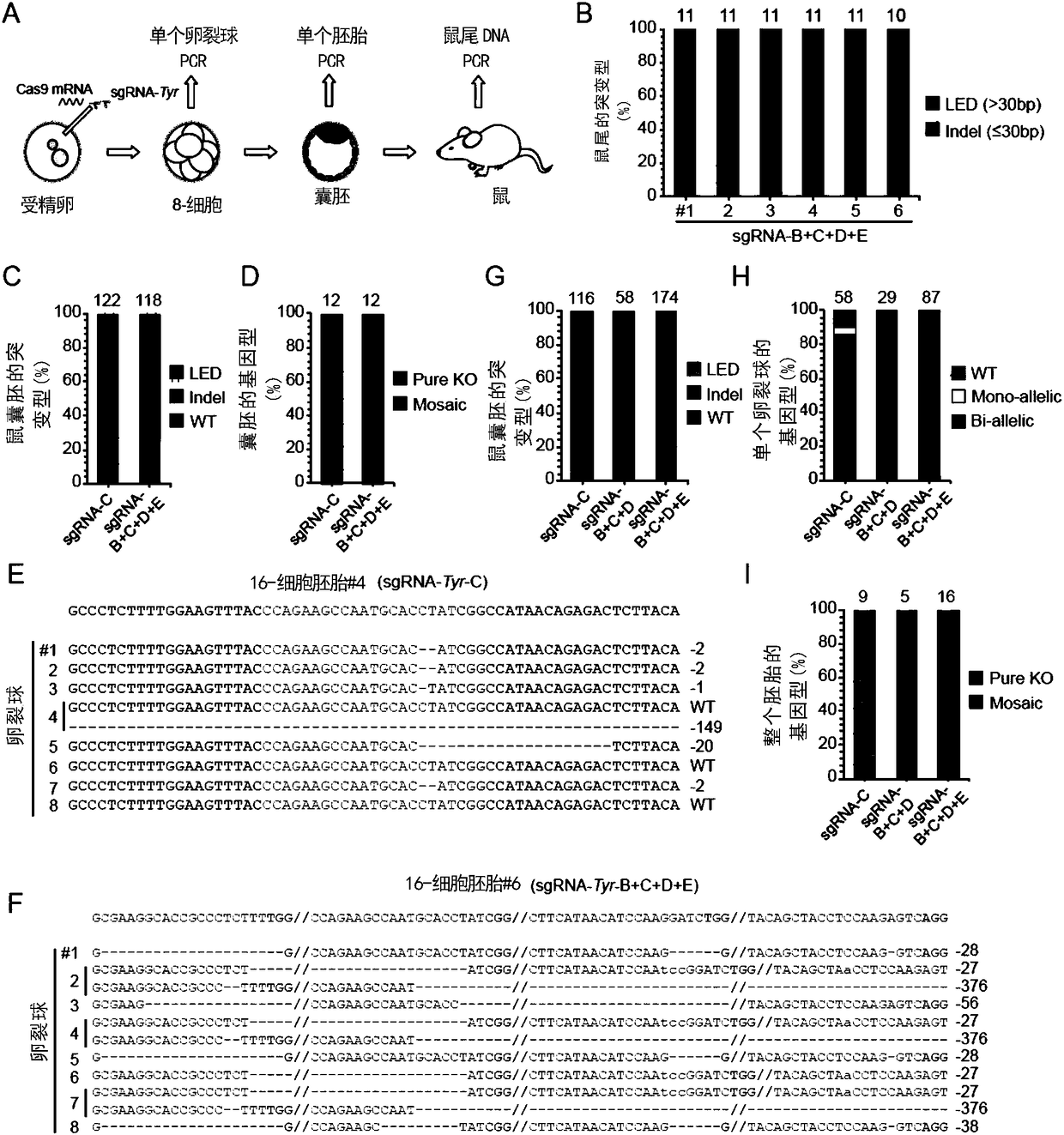
[0234] Example 3, The Mechanism of C-CRISPR Completely Deleting Genes
[0235] In order to further elucidate the mechanism of C-CRISPR, the inventors performed DNA sequence analysis on Tyr gene-edited embryos and mice. After identifying the tail tissues of 6 mice with four sgRNAs targeting Tyr (albino mice produced by sgRNA-Tyr-B+C+D+E targeting), the inventors found that the Tyr expression in the five mice The knockout was entirely due to the knockout of the entire exon, whereas the Tyr knockout in the remaining mouse was 80% due to exon knockout and 20% due to a frameshift mutation caused by an indel ( image 3 A and 3B; Figure 8 A and Figure 8 B).
[0236] Considering that the tail tissue may not be representative, the inventors also performed whole blastocyst detection on the single sgRNA group (group I) and the four sgRNA group (group IV). As expected, all group IV blastocysts (n=12) exhibited complete deletion of the wild-type Tyr allele, including 74% large exon ...
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap