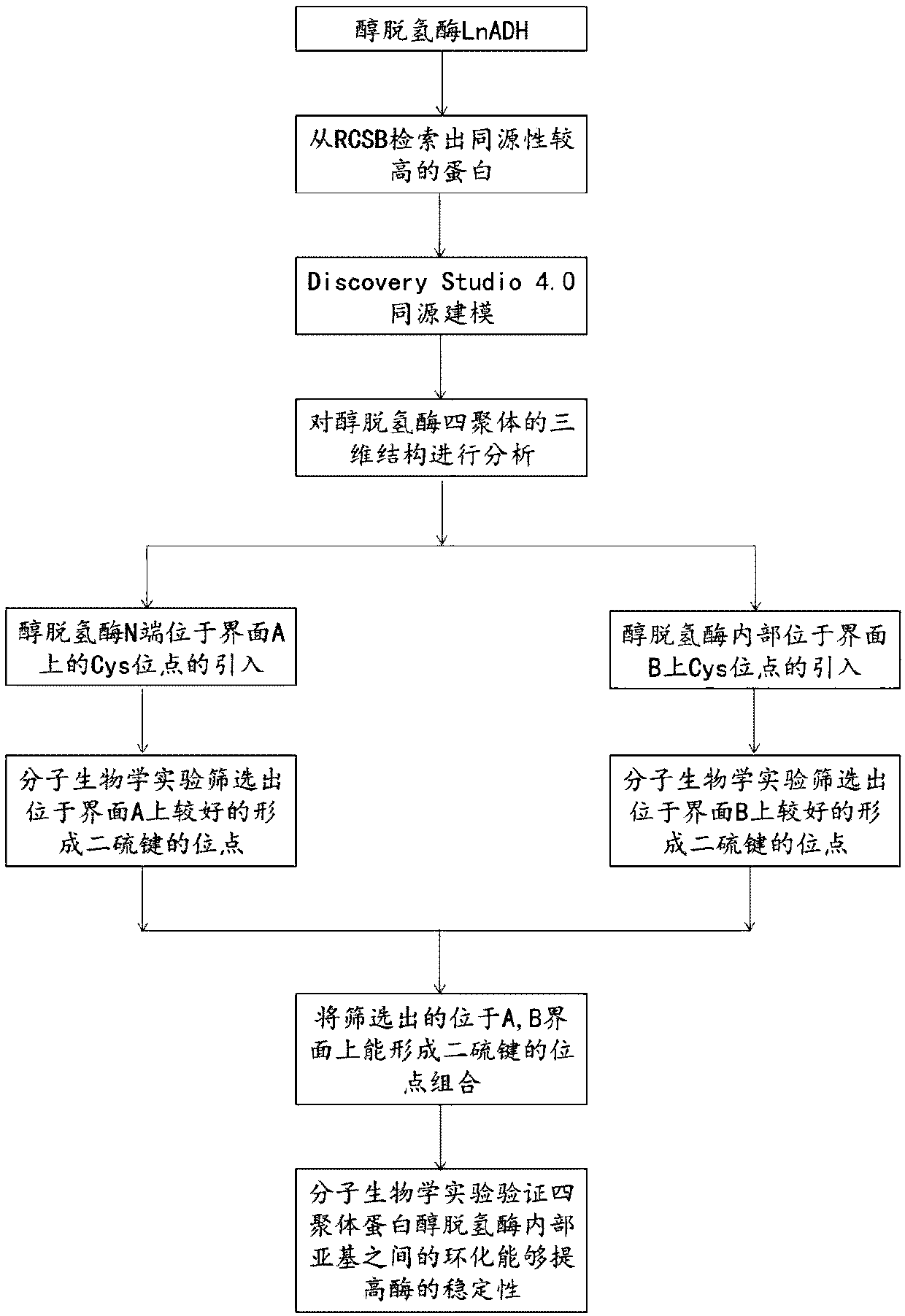
Method for increasing thermal stability of polymer protein and alcohol dehydrogenase with increased thermal stability
A thermostable, alcohol dehydrogenase technology, applied in the field of enzyme engineering, can solve problems such as low protein stability and inactivation, and achieve the effects of improving stability, avoiding cumbersome processes and broad market prospects.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
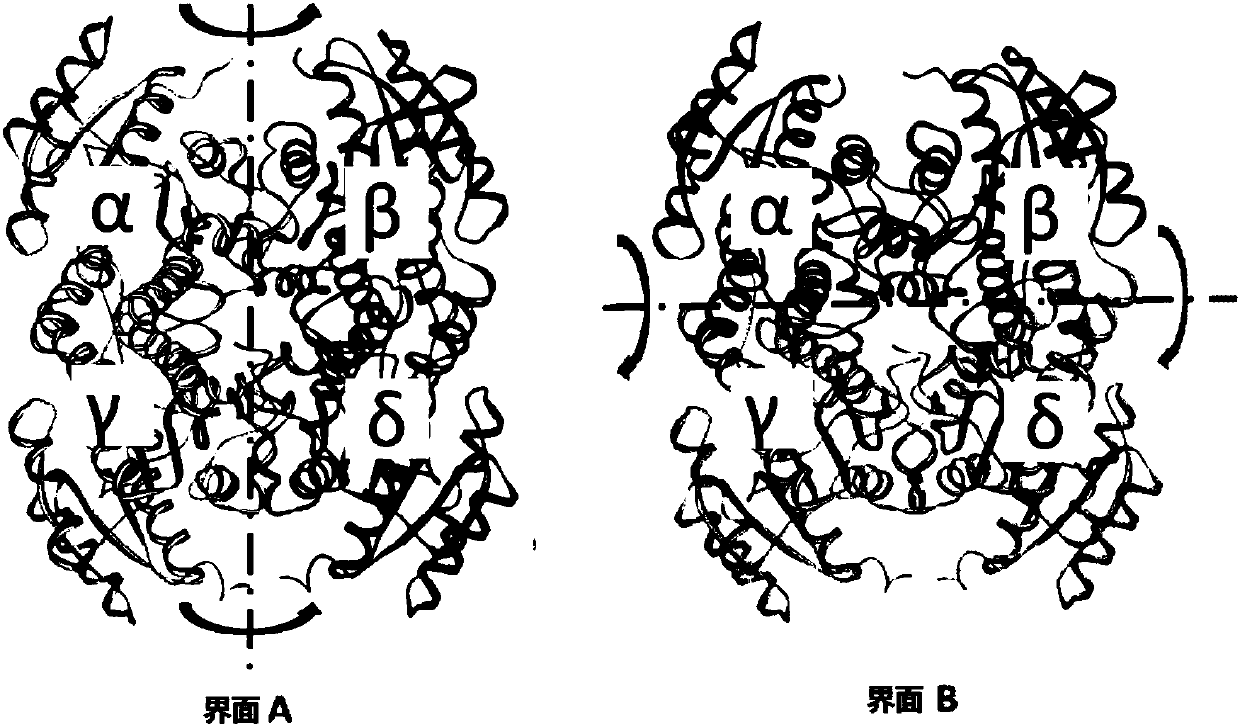
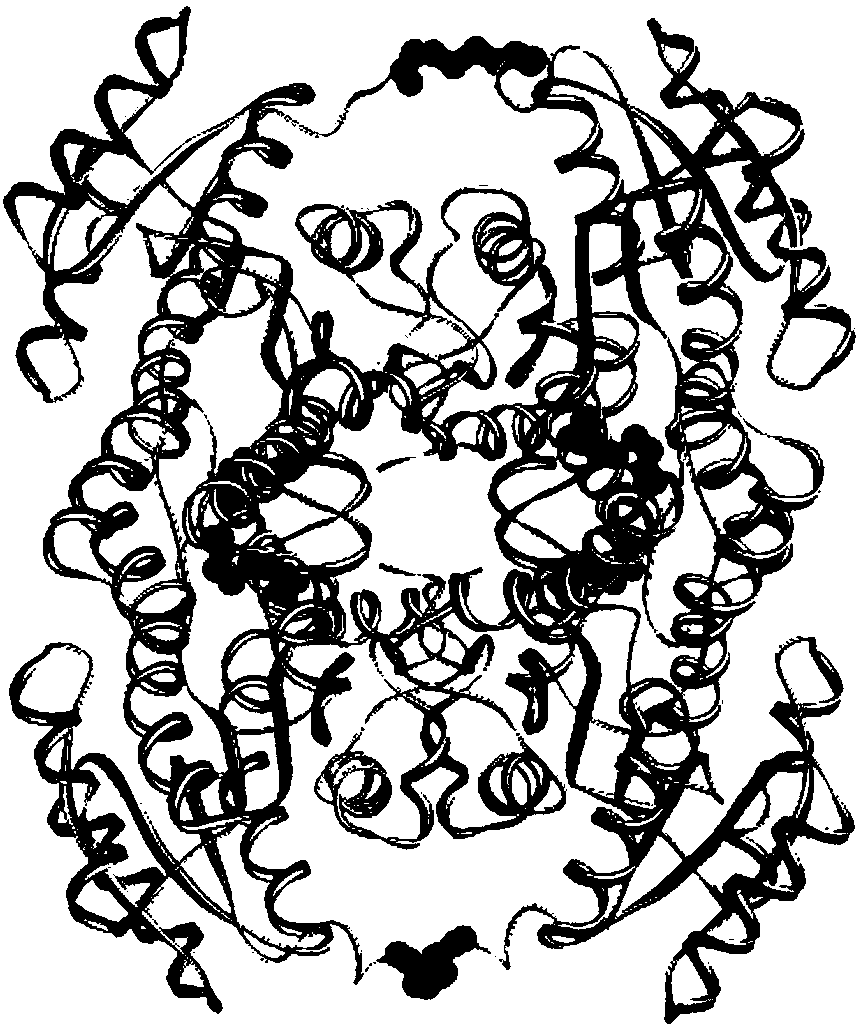
[0044] This embodiment is used to illustrate the above steps (1) by finding a protein with higher homology with alcohol dehydrogenase LnADH (SEQ ID NO.1) in the PDB database, and then using it as a template to perform homology modeling; by The protein structure was analyzed to find out the mutation site located at the N-terminal of the tetrameric protein LnADH that can form a disulfide bond on interface A. The specific method is as follows: by searching the protein database RCSB database, after blasting comparison, find the protein 4URE (PDB number, 49% homology) with high homology to the alcohol dehydrogenase, use the software Discovery Studio 4.0 to use 4URE as a template, Homology modeling of this alcohol dehydrogenase. Through the analysis of its structure, it is found that the N-terminal distance of the two subunits on the A interface is very close, only distance. Then several ways of inserting Cys at the N-terminus were devised: a. CysGlyGlySerGly amino acid sequence ...
Embodiment 2
[0046]This example is used to illustrate the above step (2) by using protein disulfide bond prediction software to find out the mutation site capable of forming a disulfide bond on the interface B of the tetrameric protein alcohol dehydrogenase LnADH. The specific method is: calculate and predict the possible disulfide bond introduction sites through the "site-directed mutagenesis" online program of the DSDBASE database (http: / / caps.ncbs.res.in / dsabase / / mainFrame.html). Through screening and exclusion, 6 groups of mutation sites with relatively high scores that can form disulfide bonds on interface B of the tetrameric protein were finally obtained, namely Val101Cys / Ala171Cys (V101C / A171C), Thr159Cys / Ala163Cys (T159C / A163C) , Thr159Cys / Leu167Cys (T159C / L167C), His162Cys (H162C), Ser155Cys / Asn170Cys (S155C / N170C), Thr100Cys / Glu174Cys (T100C / E174C).
Embodiment 3
[0048] This example is used to illustrate the above step (3) to construct a mutant that forms a disulfide bond on the contact interface A of the LnADH subunit of alcohol dehydrogenase and to verify its thermal stability. The specific method is as follows: design primers according to the GenBank accession number of alcohol dehydrogenase LnADH gene sequence (ERK72999.1), and obtain the alcohol dehydrogenase gene through PCR amplification. The test methods used include molecular cloning techniques such as PCR technology, DNA extraction, enzyme digestion, and enzyme linkage. Using Leifsonia aquatica ATCC 14665 genomic DNA as a template, the gene sequence of alcohol dehydrogenase LnADH was amplified, and this target gene was connected to the plasmid pET28aNdeI-HindIII site, and then transformed into Escherichia coli BL21 (DE 3 ) was expressed to determine its enzymatic activity. The specific process is as follows:
[0049] 1) The cultivation of alcohol dehydrogenase bacterial str...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com