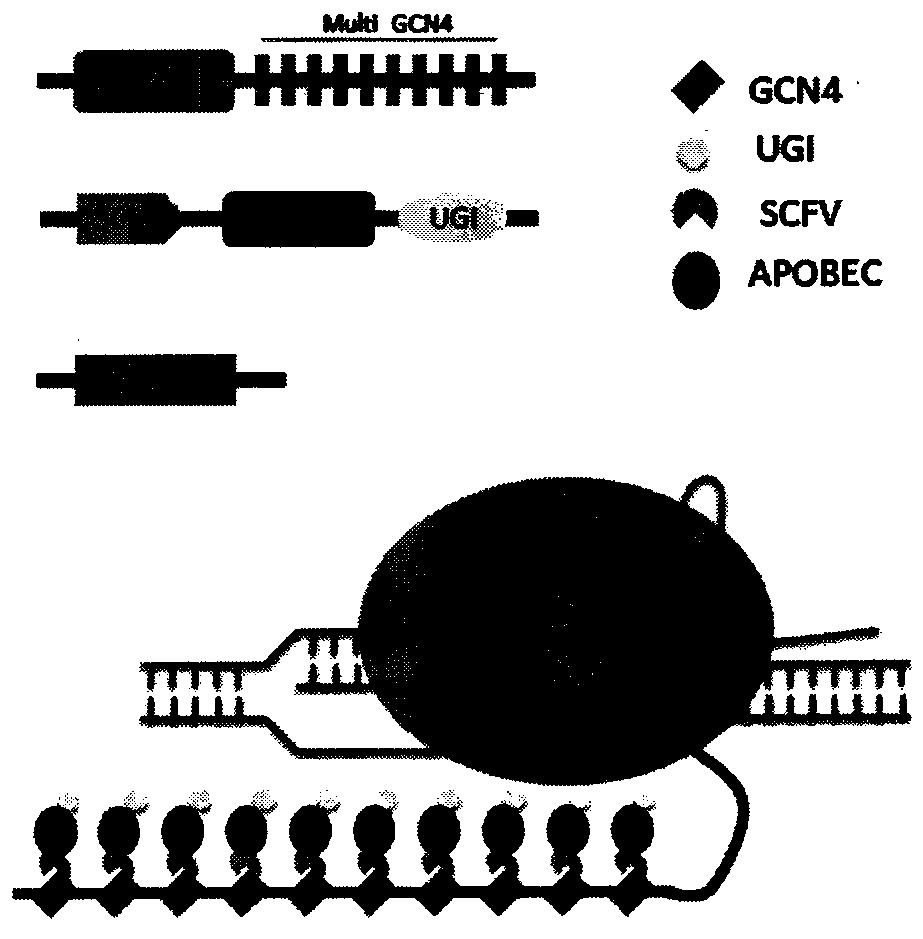
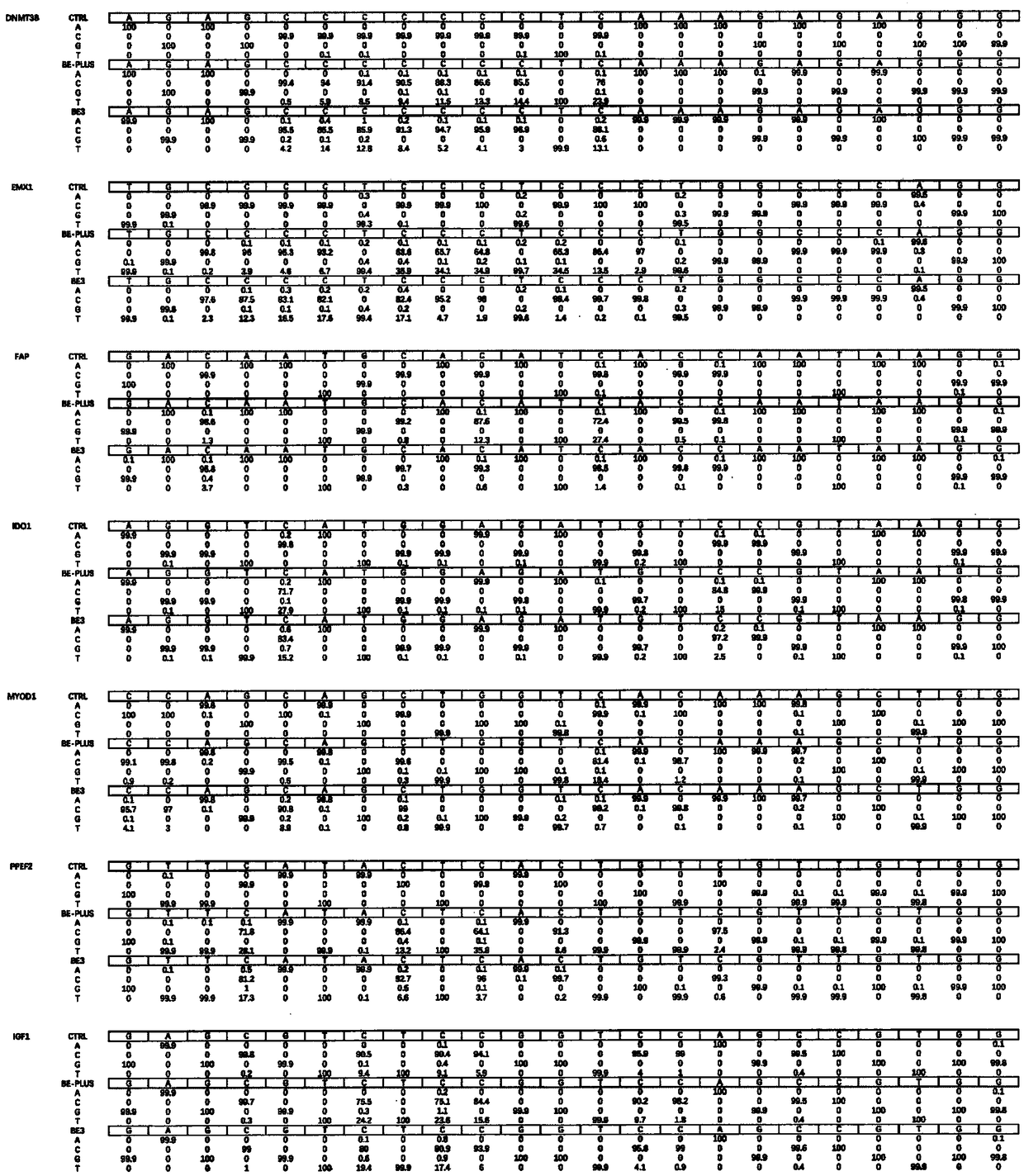
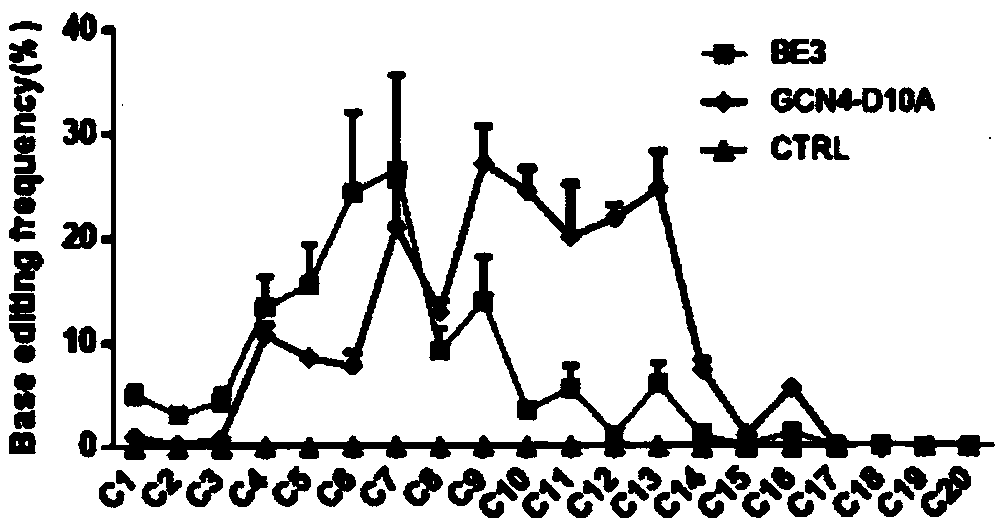
Base editing tool and application thereof
A base editing and tool technology, applied in the field of gene editing, can solve the problems of insufficient editing efficiency, small editing window, and low recognition efficiency, and achieve a large base editing range, high base editing accuracy, and editing efficiency low effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0056] 1. Construction of BE-PLUS system plasmid
[0057] 10 copies of the GCN4 sequence (10XGCN4) were synthesized by Jinweizhi Biotechnology Co., Ltd., cloned into the pUC57 vector to obtain the pUC57-10XGCN4 vector, and diluted to 10 μM as a PCR template. The forward primer was designed with NheI restriction site GCTGGCTAGCACCATGGGACCCAAGAAAAAACGCAAGGTGGAAG, and the reverse primer was designed with XbaI restriction site GTCTCTAGACACCTTGCGCTTCTTCTTGGGTCC, dissolved in water to 10 μM. A 10X GCN4 sequence fragment was amplified using Novizan High Fidelity Enzyme Kit (Vazyme, p501-d2). The amplification system and PCR reaction conditions are as follows:
[0058]
[0059]
[0060] The PCR amplification product was purified and recovered by AxyPrep PCR Clean-up Kit (Axygen, AP-PCR-500G), and 1 μg was taken, digested with NheI (NEB, R0131S) and XbaI (NEB, R0145S), and incubated at 37°C for 2h . Another 1 μg of pST1374-N-NLS-flag-linker-Cas9-D10A (Addgene #51130) vector wa...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com