A biosensor based on chain displacement and dark-state silver clusters and its application
A biosensor, strand displacement technology, applied in biochemical equipment and methods, microbial determination/inspection, etc., can solve the problems of complex operation and high cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0054] Embodiment 1 (preparation of various stock solutions)
[0055] 1. Preparation of PB buffer (PB: 20mM, pH=7.4): Accurately weigh Na 2 HPO 4 12H 2 O 7.16g in a 50mL beaker, dissolve it completely with triple distilled water and transfer it to a 100mL volumetric flask, wash the beaker and glass rod three times with ultrapure water, transfer the washing solution to the volumetric flask, dilute to 100mL, shake well 100mL 0.2M disodium hydrogen phosphate stock solution can be obtained for future use; accurately weigh the NaH 2 PO 4 2H 2 O 3.12g in a 50mL beaker, dissolve it completely with triple distilled water and transfer to a 100mL volumetric flask, wash the beaker and glass rod three times with ultrapure water, transfer the washing solution to a volumetric flask, dilute to 100mL, shake well 100mL 0.2M sodium dihydrogen phosphate stock solution can be obtained for later use; take the stock solution that has been prepared above: 19mL 0.2M NaH 2 PO 4 and 80 mL of 0.2...
Embodiment 2
[0068] Embodiment 2 (sensor construction-fluorescence detection based on strand displacement and dark-state silver clusters)
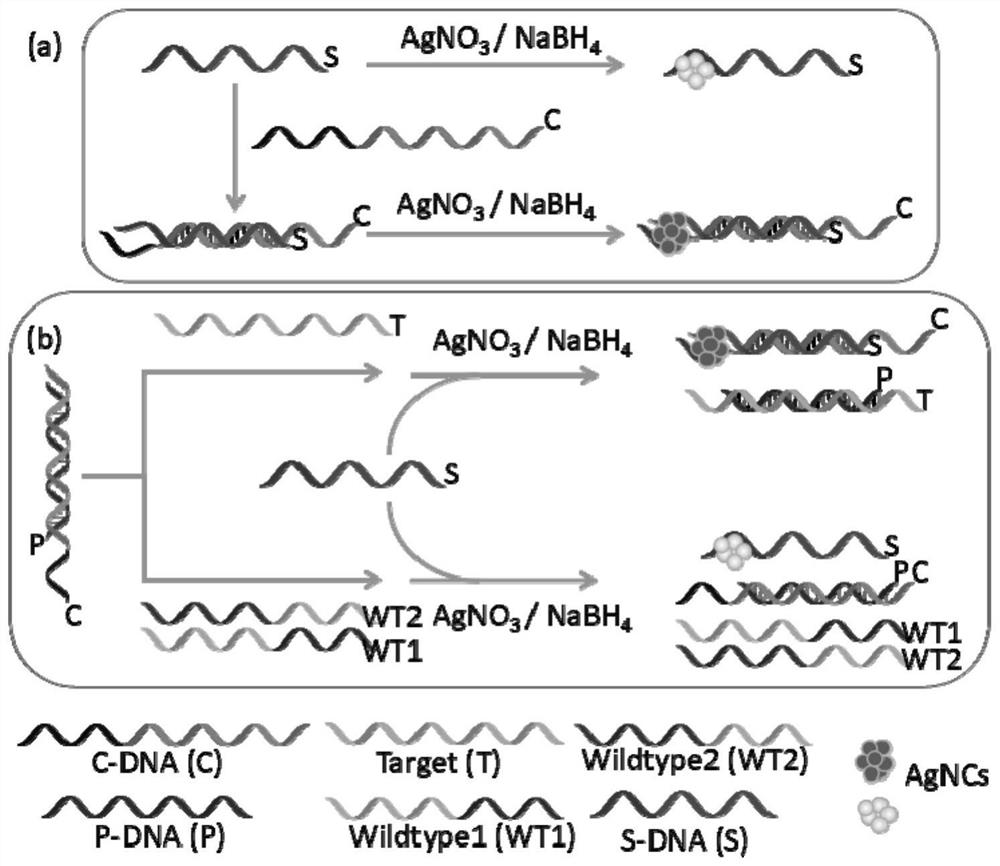
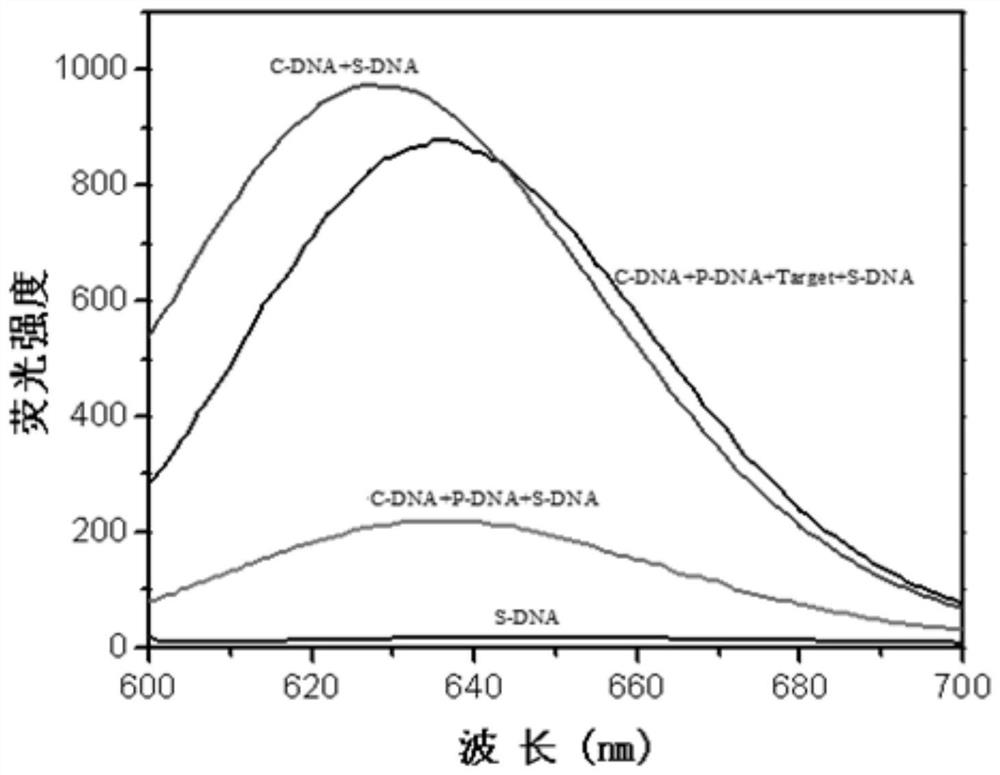
[0069] 1. Add C-DNA, C-DNA+P-DNA, C-DNA+P-DNA+Target to a 2.0ml centrifuge tube, the amount of each DNA (100μM) in each sample is 10μL, add PB buffer solution to make the system volume 200 μL. Incubate at 37°C for 1 hour;
[0070] 2. Add an appropriate amount of PB buffer solution to make the system volume 970 μL, add 10 μL of S-DNA (100 μM), 10 μL of AgNO 3 (1.8mM), magnetically shake for 1min, and incubate at room temperature for 30min in the dark;
[0071] 3. Add 10 μL fresh NaBH 4 (1.8mM), magnetically shaked for 1min, and cultured at 4°C in the dark for 5h;
[0072] 4. To detect fluorescence, the parameters are set to 10nm for the slit and 600V for the voltage. (excitation wavelength is 475nm, emission wavelength is 635nm), you can get image 3 .
Embodiment 3
[0073] Example 3 (Detection of AIMP2 Exon 2 Gene Deletion Based on Strand Displacement and Dark State Silver Cluster Sensors)
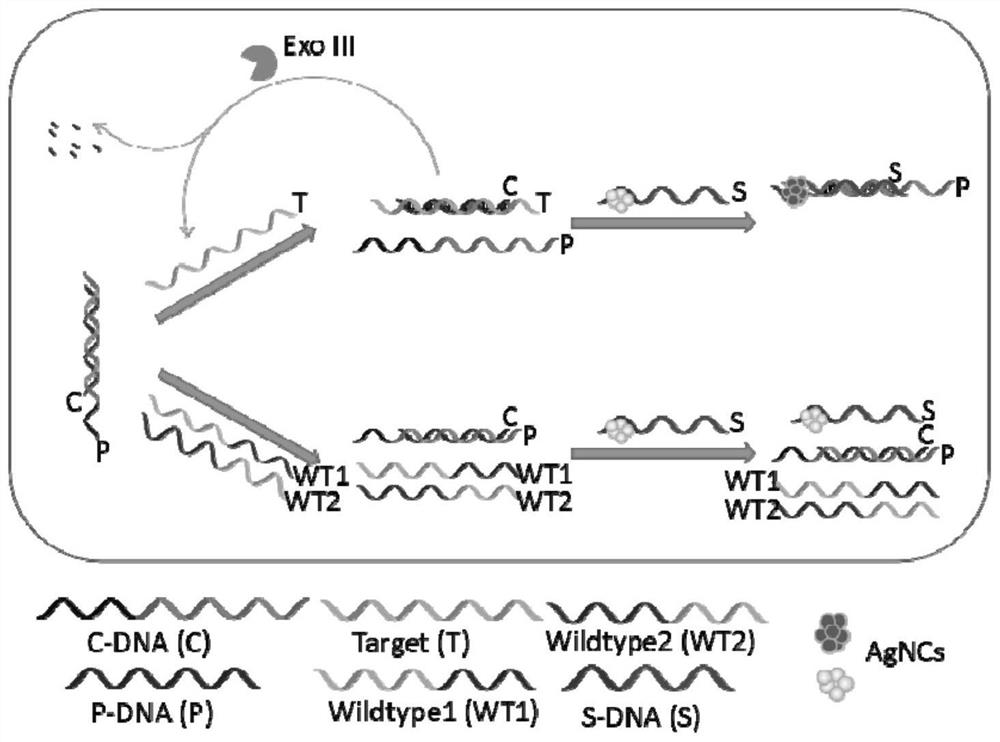
[0074] 1. Add 10 μL of C-DNA+P-DNA (both 100 μM) to a 2.0 ml centrifuge tube, add 10 μL of different concentrations of Target and different types of targets ((both 100 μM)), and add PB buffer solution to make the system volume 200 μL. Incubate at 37°C for 1 hour;
[0075] 2. Add an appropriate amount of PB buffer solution to make the system volume 970 μL, add 10 μL S-DNA (100 μM), 10 μL AgNO3 (1.8 mM), magnetically shake for 1 min, and incubate in the dark at room temperature for 30 min;
[0076] 3. Add 10 μL of freshly prepared NaBH4 (1.8 mM), shake magnetically for 1 min, and incubate at 4°C in the dark for 5 h;
[0077] 4. To detect fluorescence, the parameters are set to 10nm for the slit and 600V for the voltage. (excitation wavelength is 475nm, emission wavelength is 635nm), can obtain Fig. 4 and Figure 5 , to achieve a linear detection of ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| size | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com