A DNA methylation degree quantifying method and application
A methylation and non-methylation technology, applied in biochemical equipment and methods, microbial determination/inspection, etc., can solve the problems of limited method application scope and large sample demand, and achieve strong specificity and detection sensitivity. high effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0059] Example 1 Design and preparation of primers and probes for cg10590292
[0060] In this example, specific primers and Taqman probes were designed according to the characteristic DNA sequence of the methylation-sensitive region of the GpC island of cg10590292 and its upstream promoter region, specifically:
[0061] Methylation probe (SEQ ID NO.1):
[0062] 5'- / 6-FAM / TGGGAGAGCGGGAGAT / BHQ1 / -3';
[0063] Unmethylated probe (SEQ ID NO.2):
[0064] 5'- / HEX / TTTGGGAGAGTGGGAGATTT / BHQ1 / -3';
[0065] Upstream primer (SEQ ID NO.3):
[0066] 5'-TGTTAGTTTTTTATGGAAGTTT-3';
[0067] Downstream primer (SEQ ID NO.4):
[0068] 5'-AAACAACAAAATACTCAAA-3'.
Embodiment 2
[0069] Example 2 DNA extraction and sulfite modification
[0070] (1) Take 10mL of blood samples from the subjects, store them in Streck Cell-Free DNA BCT plasma free DNA blood collection tubes, and conduct plasma separation;
[0071] (2) ctDNA in plasma was extracted using ctDNA Extraction Kit (QIAamp Circulating Nucleic Acid Kit);
[0072] (3) Using the DNA sulfite modification kit (EZ DNA Methylation-Direct Kit), strictly follow the operation steps of the manual to treat the extracted ctDNA with sulfite, so that the unmethylated cytosine in the ctDNA is deaminated and transformed into Uracil, while methylated cytosine remains unchanged, yields sulfite-converted ctDNA.
Embodiment 3
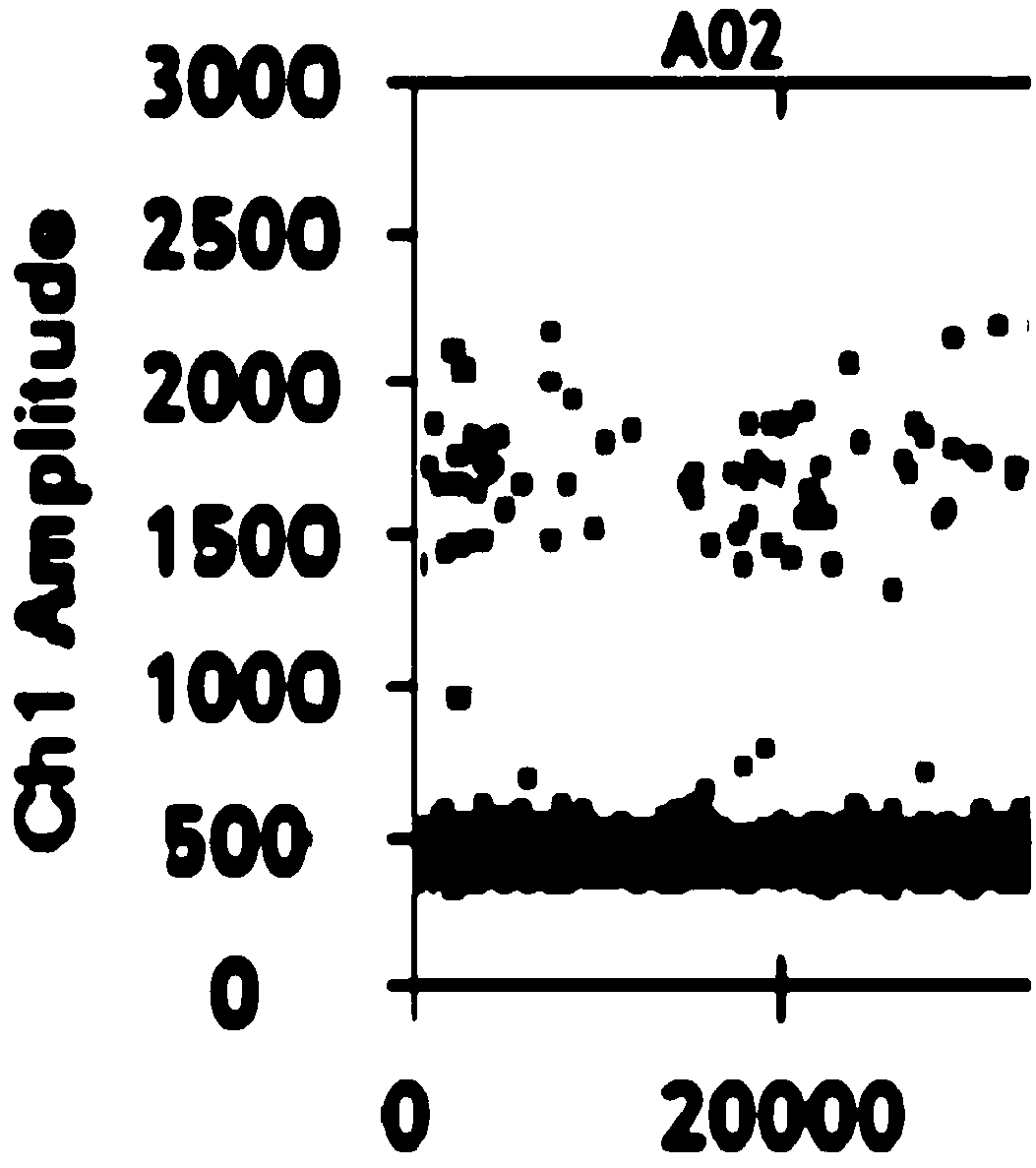
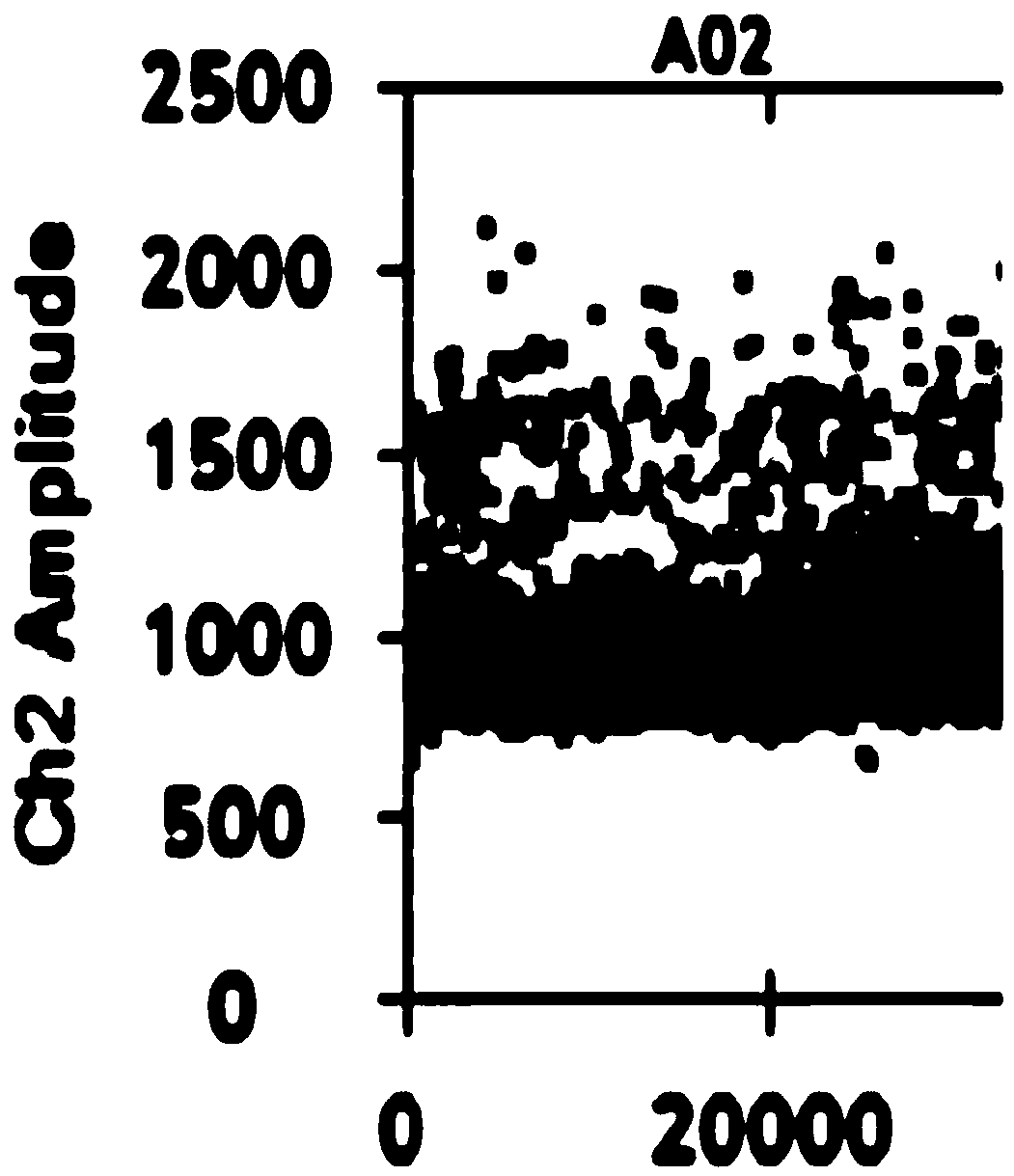
[0073] Embodiment 3 digital PCR
[0074] (1) Configure methylated PCR system and unmethylated PCR system respectively, including ctDNA template, primer pair, Taqman probe, DNA polymerase and dNTP, etc. The primer pair and Taqman probe used are shown in Table 1;
[0075] (2) Add 20 μL of sulfite-modified ctDNA samples to each of the 8 wells in the middle row of the DG8cartridge droplet generation card. Add 70 μL micro-droplet generating oil;
[0076] (3) Mix evenly by inverting, centrifuge to remove air bubbles, put into the micro-droplet generator, and generate micro-droplets;
[0077] (4) Transfer the droplet to a 96-well plate, and perform PCR amplification after sealing the film. The amplification conditions are: 95°C, 10min, 1 cycle; 95°C, 30s, 60°C, 60s, 40 cycles; 98°C, 60s, 40 cycles; 10min;
[0078] (5) After assembling the 96-well plate that completed the PCR reaction, put it into the droplet reader smoothly, perform absolute quantification of the DNA in the two PC...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com