Specific sgRNA for target knocking out human OC-2 genes and application
An OC-2, specific technology, applied in the fields of molecular biology and biomedicine, can solve problems such as difficulties and off-targets, and achieve the effect of reducing proliferation and good application prospects.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0057] Example 1 CRISPR / Cas9 Gene Knockout Vector Construction
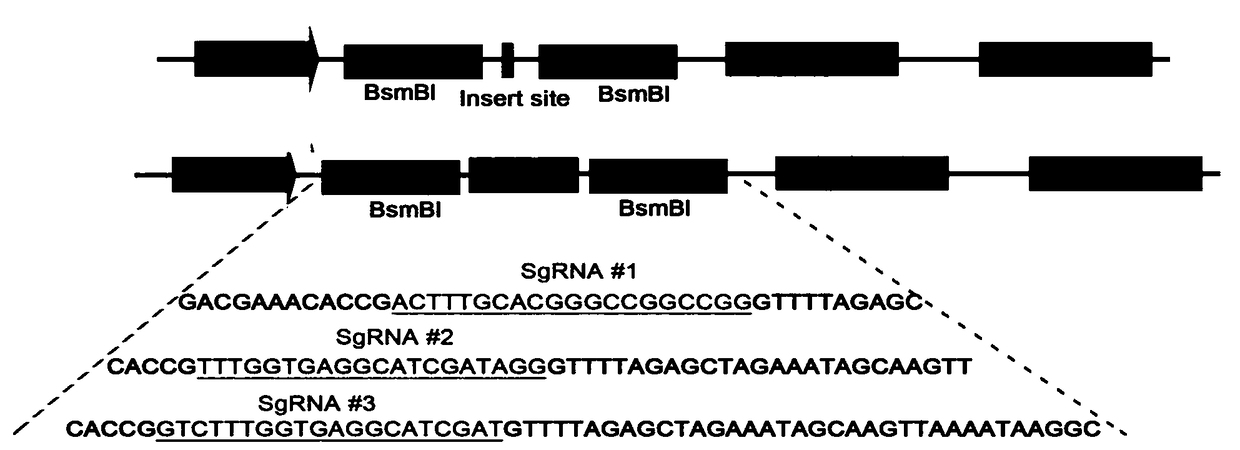
[0058] 1. Selection and design of sgRNA targeting human OC-2 gene
[0059] The sequence of the CDS region of the OC-2 gene was analyzed in the NCBI database to determine the site to be knocked out, and finally the knockout site was determined to be exon 1 of the OC-2 gene. Through the sgRNA online design website (http: / / crispr.mit.edu / ), three high-scoring specific sgRNAs targeting exon 1 of the ONECUT2 gene were designed, and the off-target efficiency of the sgRNAs was evaluated.
[0060] The sequence of exon 1 is as follows (SEQ ID NO.1):
[0061] ATGAAGGCTGCCTACACCGCCTATCGATGCCTCACCAAAGACCTAGAAGGCTGCGCCATGAACCCGGAGCTGACAATGGAAAGTCTGGGCACTTTGCACGGGCCGGCCGGCGGCGGCAGTGGCGGGGGCGGCGGCGGGGCGGCGGGGGCGGCGGCGGGGGCCCGGGCCATGAGCAGGAGCTGCTGGCACGCCCCCCC
[0062] sgRNA#1 (SEQ ID NO.2):
[0063] ACTTTGCACGGGCCGGCCGG
[0064] sgRNA#2 (SEQ ID NO.3):
[0065] TTTGGTGAGGCATCGATAGG
[0066] sgRNA#3 (SEQ ID NO.4):
[0067] ...
Embodiment 2
[0100] Example 2 Cell Culture
[0101] Human ovarian cancer cell lines SKOV3, CAOV3, and human embryonic kidney epithelial cells (293T) were purchased from the ATCC Culture Center. The cells were cultured with DMEM medium (Gibco Company) containing 10% FBS, penicillin and streptomycin (Invitrogen Company) were added to the medium, and the final concentrations were 100 U / mL and 100 μg / mL, respectively, and cultured at 37°C Carbon dioxide incubator.
Embodiment 3
[0102] Example 3 lentiviral packaging
[0103] Human 293T cells in the logarithmic growth phase were digested with trypsin and finally prepared into a single cell suspension. The cells were counted (repeated three times), and the cell concentration was adjusted to 5×10 with DMEM complete medium 4 individual / mL. 293T cells were seeded in 100mm cell culture dishes and cultured by adding 7mL DMEM complete medium.
[0104] The day before transfection, the 293T cell culture medium was replaced with DMEM basal medium (without adding double antibody) for starvation culture. Core plasmid (LentiCRISPRv2 recombinant plasmid) 4 μg, psPAX2 (purchased from Addgene) 3 μg, pMD2G (purchased from Addgene) 1 μg, transfection reagent (FuGENE6) 24 μL supplemented with DMEM basal medium (without adding double antibody) to prepare a total volume of 200 μL transfection After incubating for 30 minutes in a cell culture incubator at 37°C, add it dropwise to a 293T cell culture dish that is almost c...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com