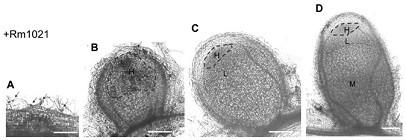
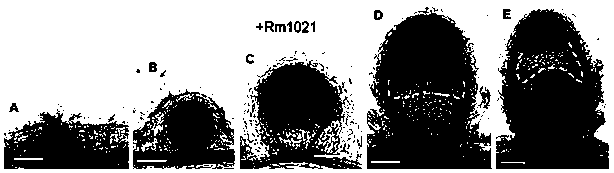
Promoter report gene for detecting superoxide radical ions in nodules and application thereof
A superoxide ion and reporter gene technology, applied in the direction of oxidoreductase, using vectors to introduce foreign genetic material, enzymes, etc., can solve the problem of unclear distribution pattern of ROS
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0018] Embodiment one: build sodB Promoter- uidA gene fusion plasmid
[0019] Using Sinorhizobium meliloti DNA as a template, each with a restriction enzyme site Pst I and Bam HI upstream and downstream primers , Cloned by polymerase chain reaction (PCR) sodB For the promoter sequence of the gene, refer to the DNA sequence shown in SEQ ID NO: 1 in the sequence listing. The PCR product was digested by restriction endonuclease and uidA Gene plasmid pRG960, and then use T4 ligase to connect the PCR product to the plasmid. Finally, the constructed plasmid was transformed into competent Escherichia coli DH5α by heat shock method.
Embodiment 2
[0020] Embodiment two: construction plasmid is transferred into S. meliloti
[0021] Transfer into mutant strains using the three-parent conjugation transfer experiment:
[0022] a. Inoculate Rhizobia (mutant strain), Escherichia coli (constructed fusion plasmid) and helper vector MT616, and culture at 28 ℃ and 37 ℃ until OD 600 is about 1;
[0023] b. Take 1000 µL of each of the above strains in a centrifuge tube, centrifuge at 8000 rpm for 1 min, remove the supernatant and retain the precipitate;
[0024] c. 1 ml of 0.85% sterilized normal saline to suspend and precipitate once to remove antibiotics;
[0025] d. Centrifuge at 8000 rpm for 1 min, remove the supernatant and retain the precipitate;
[0026] e. Use 300 µL of freshly prepared liquid LB medium to suspend the Rhizobia, E. coli and MT616 pellets;
[0027] f. Spot 5 µL of each of the three bacterial solutions on the same position on the non-resistant LB plate, and mix carefully to prevent the bacterial solution f...
Embodiment 3
[0045] Embodiment three: Inoculate the host plant Medicago sativa into the rhizobia with the fusion plasmid
[0046] (1) Germination of alfalfa seeds:
[0047] 1) Add alfalfa seeds to a sterile small triangular flask, add 75% ethanol and gently shake the triangular flask to make the seed
[0048] The seeds were fully exposed to ethanol for 5 min. Pour off all ethanol.
[0049] 2) Quickly pour a large amount of sterile water into the triangular flask and shake it quickly. Then pour out the water in the triangular flask,
[0050] Then repeat the water change three times to wash off the residual ethanol thoroughly.
[0051]3) Add 25% antiformin (sodium hypochlorite) solution to the Erlenmeyer flask. Shake gently for 15 min, then
[0052] After washing with water for 3-5 times, change the water every 10 minutes, repeat 5 times, in order to fully remove the seeds
[0053] Sodium hypochlorite residue on the surface.
[0054] 4) Add 20 mL of sterile water to the Erlenmeyer fl...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com