Aptamer for specifically recognizing GTONNV (Guangxi trachinotus ovatus nervous necrosis virus) and application of aptamer
A nucleic acid aptamer, a technology for oval pomfret, which is applied in the direction of microbial determination/test, chemical library, combinatorial chemistry, etc., can solve the problems of inability to detect and diagnose oval pomfret-derived nerve necrosis virus, and achieves convenient in vitro Chemical synthesis, high affinity and specificity, good reproducibility
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] The preparation method of embodiment 1ssDNA nucleic acid aptamer is as follows:
[0035] Step 1: Synthesize the single-stranded DNA library and primers shown in the following sequences:
[0036] Random library Library50:
[0037] 5'-GTCTGAAGTAGACGCAGGAG(50N)AGTCACACCTGAGTAAGCGT
[0038] 5' primer: 5'-FAM-GTCTGAAGTAGACGCAGGAG-3';
[0039] 3' Primer: 5'-Biotin-ACGCTTACTCAGGTGTGACT-3';
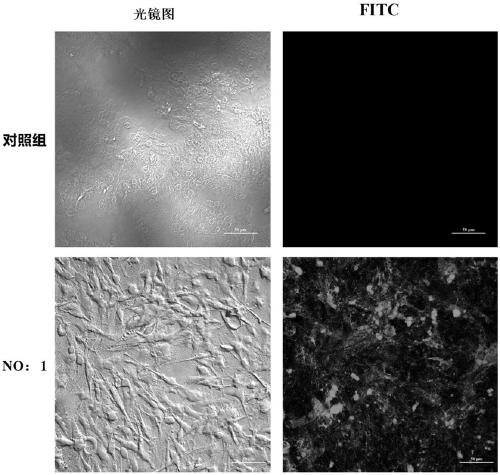
[0040] Step 2: Dissolve 10 nmol of the above random library in 500 μl PBS, place in a constant temperature water bath at 92°C for 5 minutes, then quickly ice-bath for 10 minutes, take the treated random library and incubate GTONNV-infected cells on ice for 1 hour; wait for incubation After the combination is completed, centrifuge and remove the supernatant, take 10mL of PBS to wash the GTONNV infected cells, and in a constant temperature water bath at 92°C for 10min, centrifuge at 12000g for 1-20min, and collect the supernatant, which is To specifically recognize the ssDNA nucleic acid...
Embodiment 2
[0052] The preparation method of embodiment 2 ssDNA nucleic acid aptamer is as follows:
[0053] Step 1: Synthesize the single-stranded DNA library and primers shown in the following sequences:
[0054] Random library Library50:
[0055] 5'-GTCTGAAGTAGACGCAGGAG(50N)AGTCACACCTGAGTAAGCGT
[0056] 5' primer: 5'-FAM-GTCTGAAGTAGACGCAGGAG-3';
[0057] 3' Primer: 5'-Biotin-ACGCTTACTCAGGTGTGACT-3';
[0058] Step 2: Dissolve 10 nmol of the above random library in 500 μl PBS, place in a constant temperature water bath at 92°C for 5 minutes, then quickly ice-bath for 10 minutes, take the treated random library and incubate GTONNV-infected cells on ice for 1 hour; wait for incubation After the combination is completed, centrifuge and remove the supernatant, take 10mL of PBS to wash the GTONNV infected cells, and in a constant temperature water bath at 92°C for 10min, centrifuge at 12000g for 1-20min, and collect the supernatant, which is To specifically recognize the ssDNA nucleic aci...
Embodiment 3
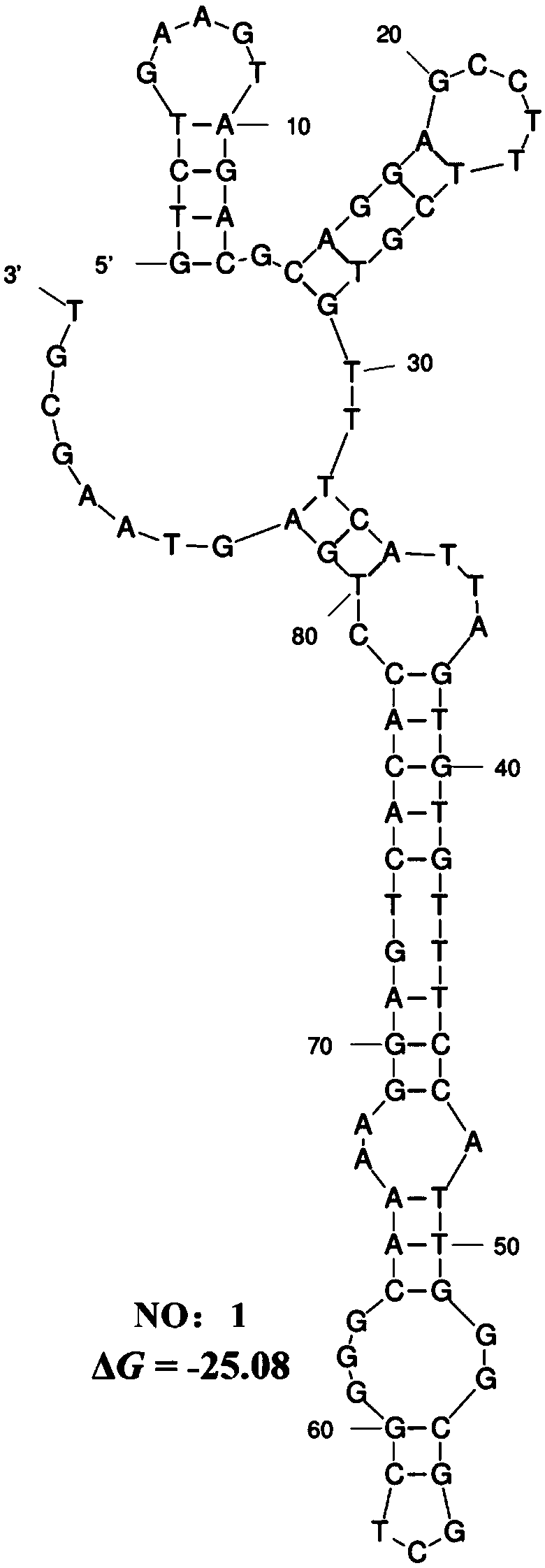
[0082] The secondary structure of the nucleic acid aptamer was predicted online by MFOLD software.
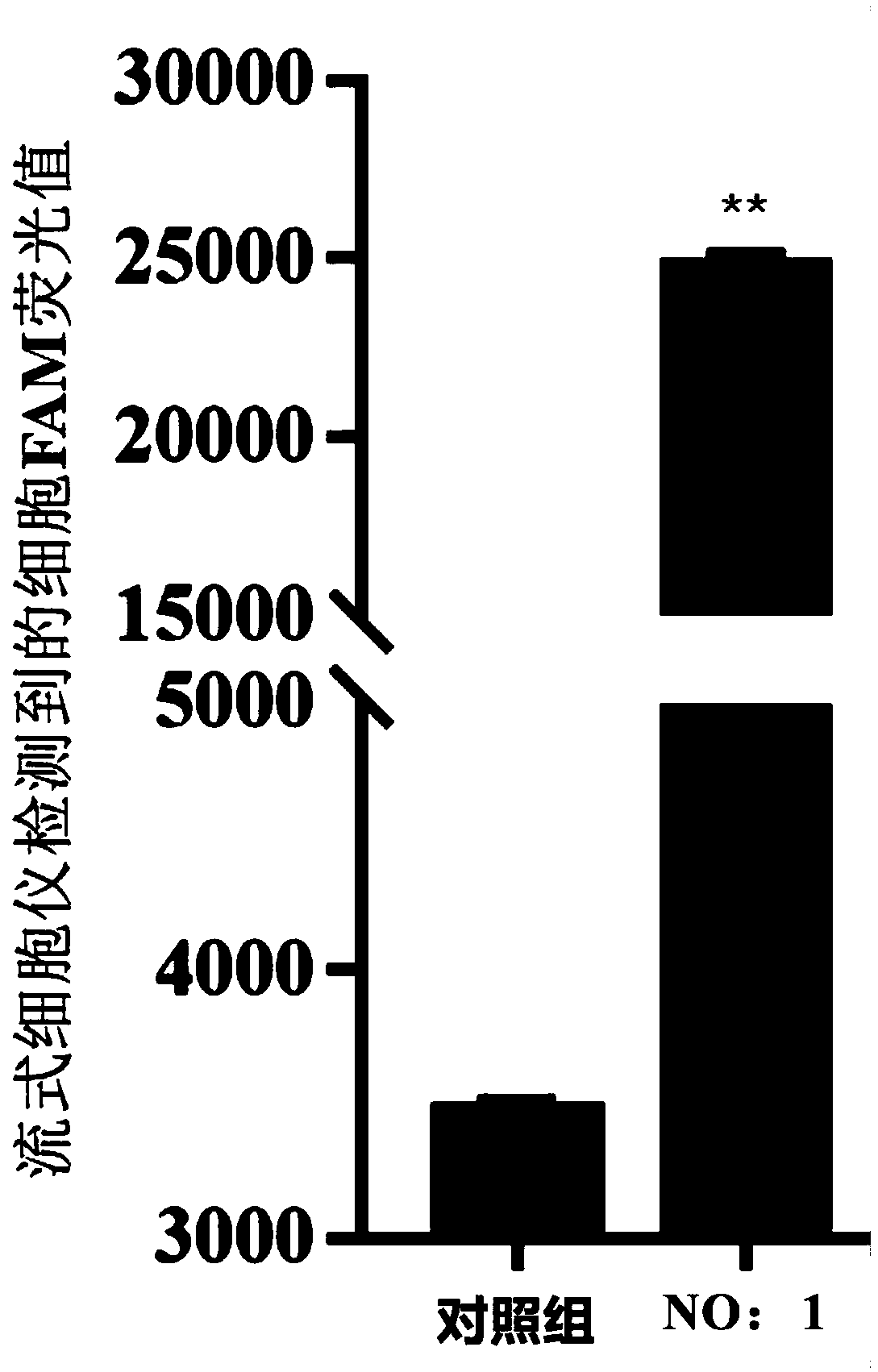
[0083] The secondary structure prediction results of the nucleic acid aptamer of SEQ ID NO:1 are as follows image 3 As shown, the nucleic acid aptamer forms a special stem-loop structure and hairpin structure.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com