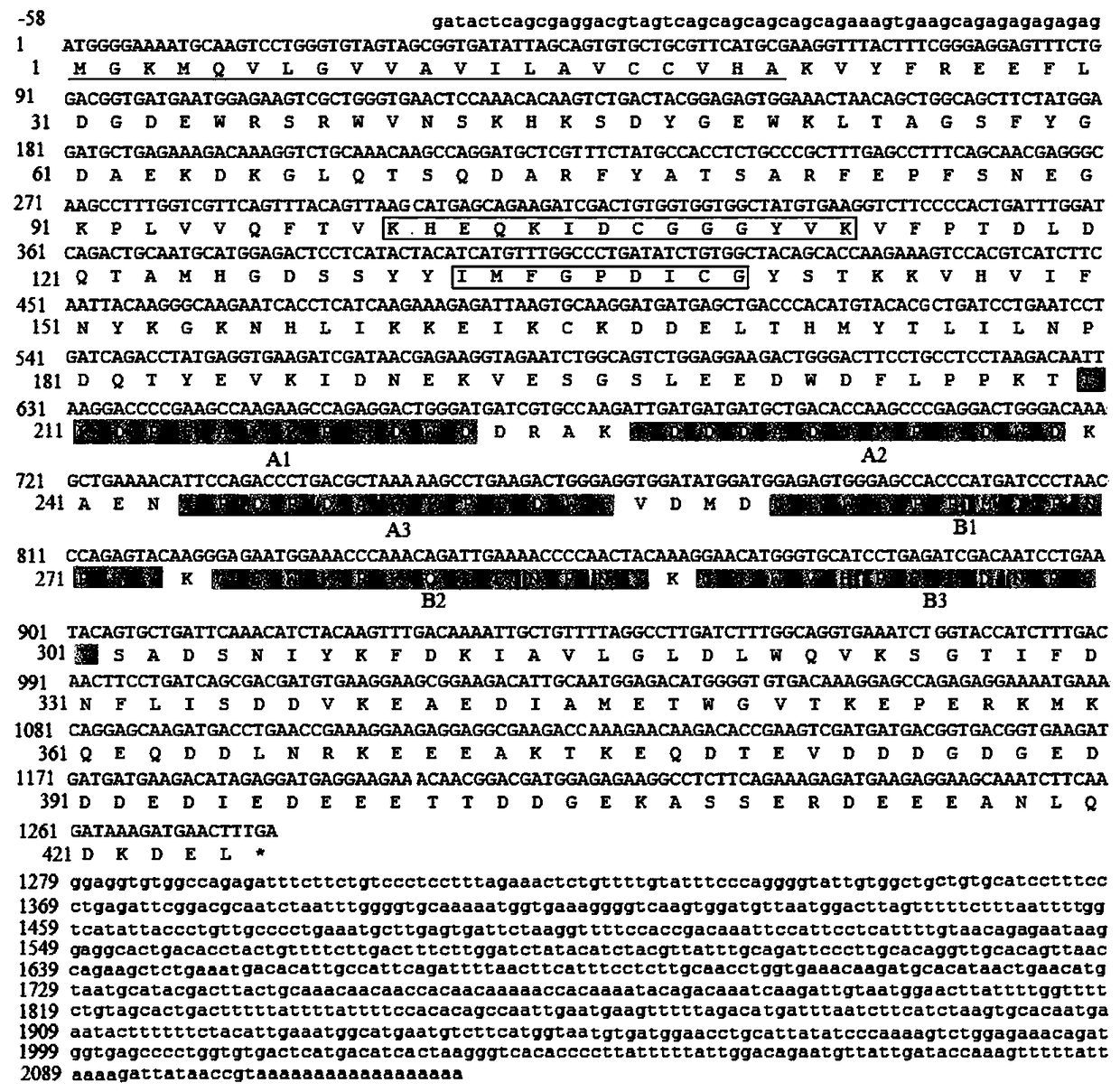
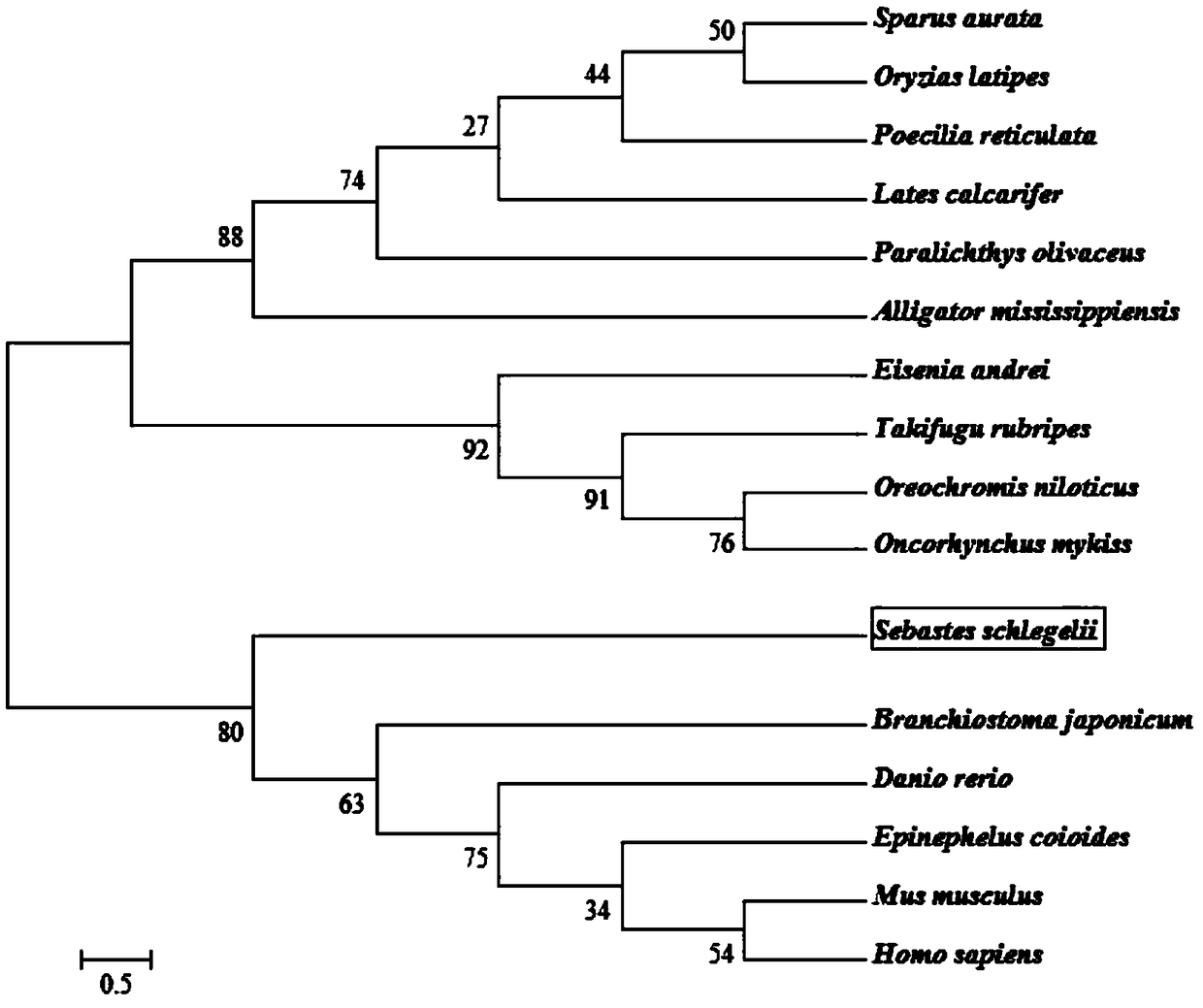
Sebastes schlegeli calreticulin gene and application
A calreticulin and gene technology, applied in application, genetic engineering, plant genetic improvement and other directions, can solve problems such as unreported, and achieve the effects of low cost, strong specificity and simple operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Example 1 Extraction of Sebastes schlegeli DNA
[0034] 1. Extraction of kidney RNA:
[0035] RNApreppure Tissue Kit (TIANGEN) kit to extract total RNA, the specific steps are as follows:
[0036] (1) Homogenization treatment: add 400μl lysis solution RL to about 10mg of tissue, grind with a pestle, and add 600μl RNase-Free ddH after grinding. 2 O and 10μl proteinase K, mix well and then bathe in 56℃ water for 15min, 12000rpm, 5min, take the supernatant and transfer to RNase-free sterile 1.5ml centrifuge tube.
[0037] (2) Add absolute ethanol: add 0.5 times the volume of the supernatant to a 1.5ml centrifuge tube containing the supernatant, mix and transfer to the adsorption column, put it in a centrifuge, centrifuge at 12000rpm for 1min, Discard the waste liquid.
[0038] (3) Add protein-removed solution: pipet 350μl protein-removed solution RW1 and add it to the adsorption column, centrifuge at 12000rpm for 1min, and discard the waste solution.
Embodiment 2
[0052] Example 2 Homologous cloning of Xu's Pingyu CRT (SsCRT) gene
[0053] According to the homologous gene cloning method, referring to the CRT gene sequences of other fishes reported in GenBank, the conservative sequences of the CRT gene family were selected by sequence alignment, and a pair of degenerate primers CRT-F and CRT-R were designed (Table 1), All primers were synthesized by Beijing Kinco Biotechnology Co., Ltd., and PCR amplification was performed using cDNA of Xu's Pingyu kidney as a template.
[0054] Table 1 Scallop CRT gene homologous cloning primers
[0055]
[0056]
[0057] (1) PCR reaction system and reaction conditions:
[0058]
[0059]
[0060] (2) DNA fragment recovery
[0061] After the PCR products are separated and detected by 1.2% agarose gel electrophoresis, they are recovered and purified by DNA gel extraction kit. The specific steps are as follows:
[0062] ①Cut the gel: Cut the gel containing the target DNA band under the long-wave 365nm UV lamp and pu...
Embodiment 3
[0071] Example 3 RACE (Rapid-amplification of cDNA ends) technology to clone the full-length sequence of CRT gene
[0072] According to the obtained homologous conservative sequence of the Calreticulin gene of Xu's Pingyu, the specific primers required for 5'and 3'RACE were designed respectively (see Table 2.2), and nested PCR was performed with the adapter primers (see Table 2.2) (Nested-PCR) reaction to amplify the 5'end sequence and 3'end sequence of the target gene. Follow SMART TM RACE cDNA amplification kit operating instructions, respectively synthesize 5’-RACE-Ready-cDNA and 3’-RACE-Ready-cDNA as templates for 5’ and 3’ PCR amplification.
[0073] (1) Synthesis of the first strand of RACE-Ready cDNA template:
[0074]
[0075] ②Mix, centrifuge, heat denature at 70℃ for 2min, ice bath for 2min, and centrifuge.
[0076] ③Add in each tube:
[0077]
[0078] ③In the PCR machine, get the cDNA template at the 5'and 3'ends at 42°C, 1.5h, 72°C, 7min
[0079] (2) Perform nested PCR w...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com