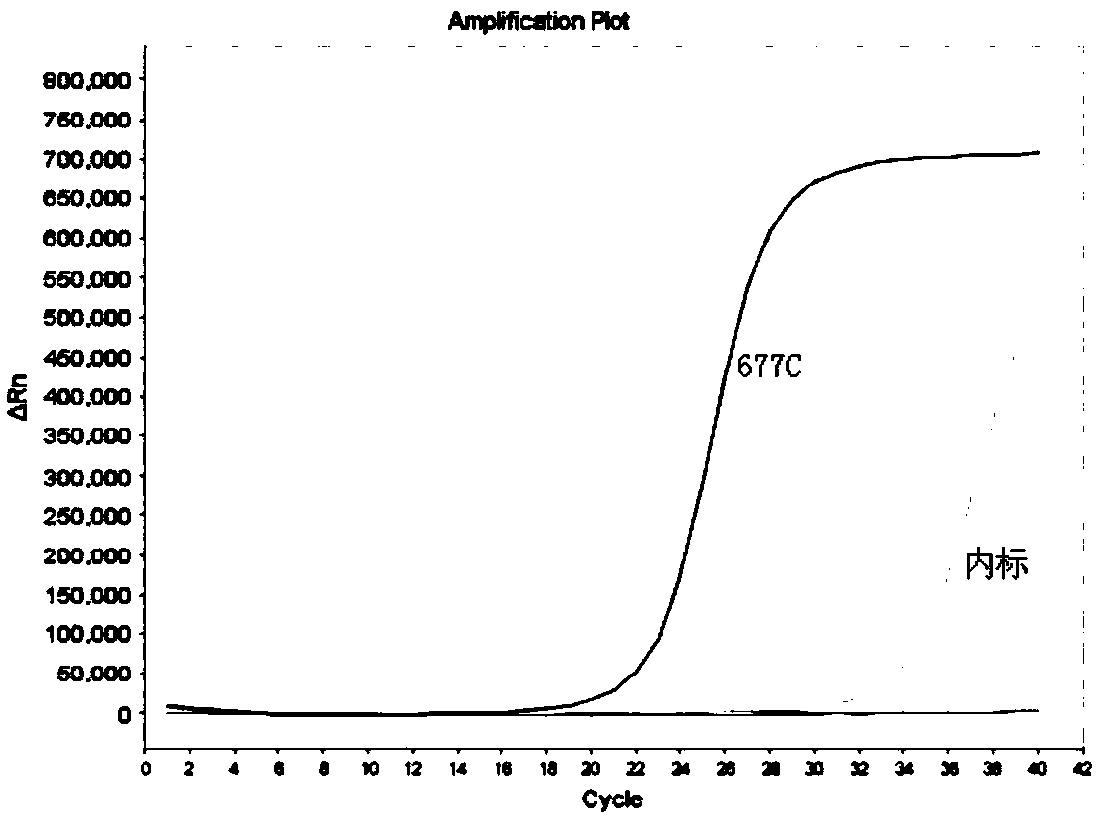
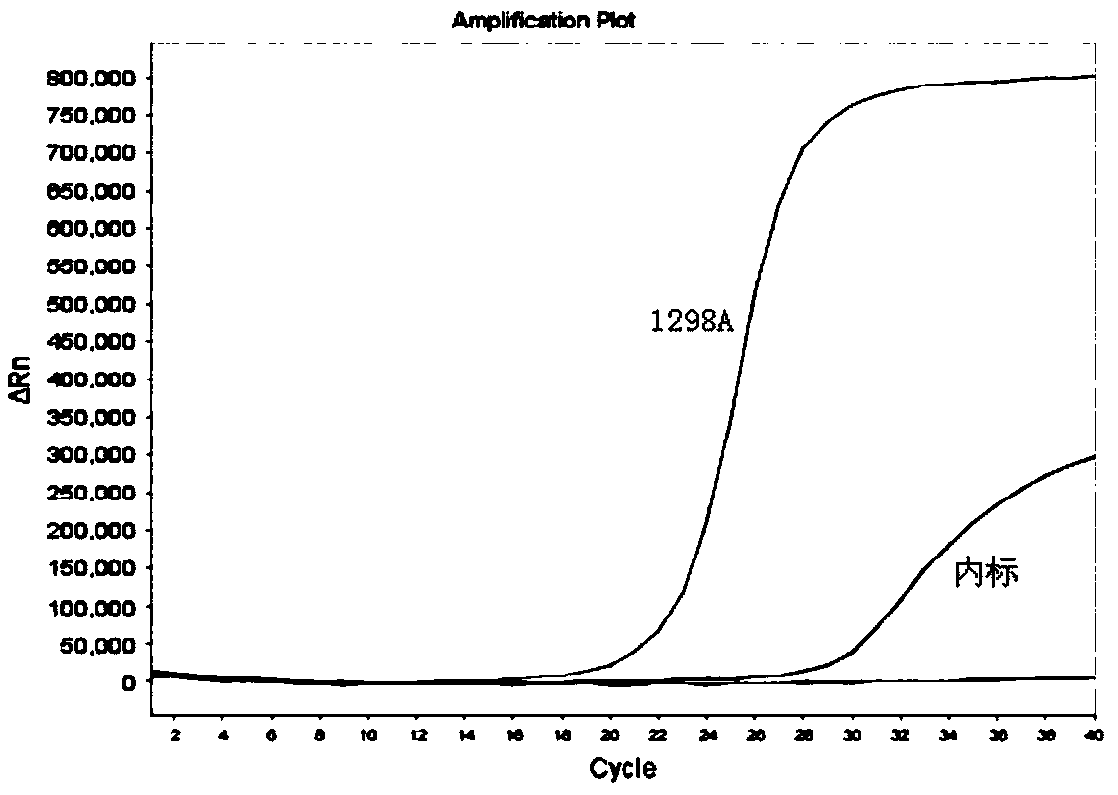
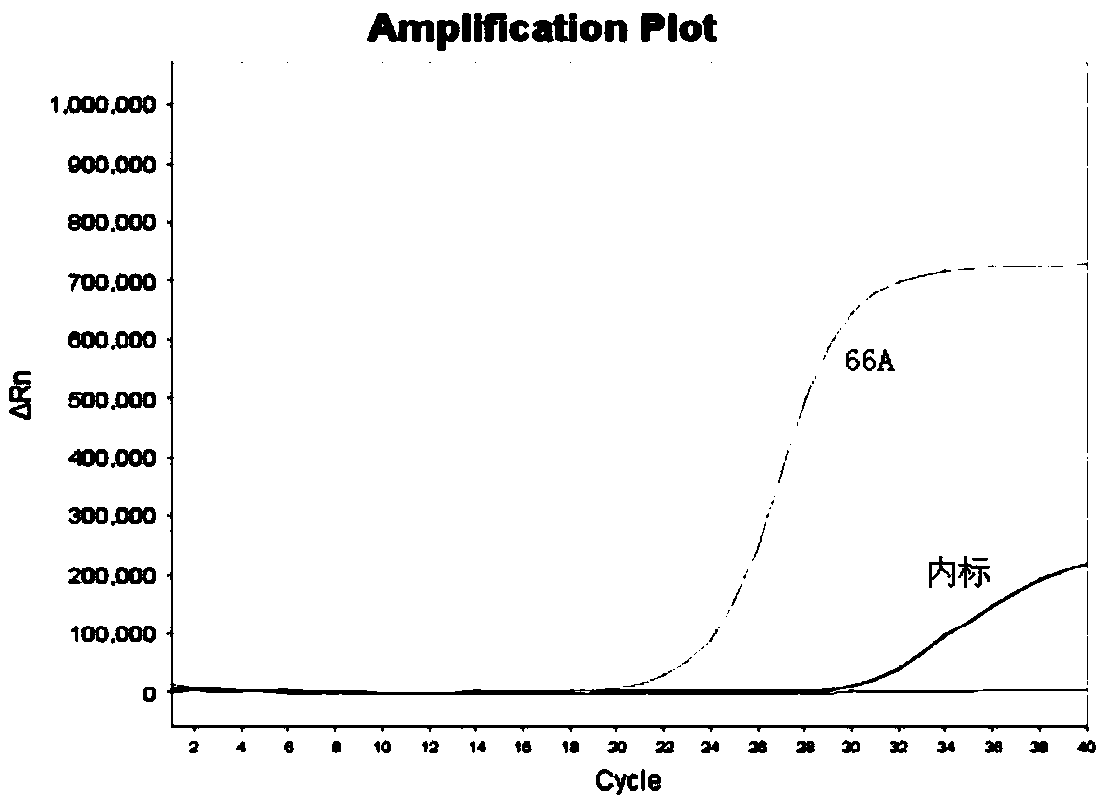
Kit for detecting genotyping of folic acid metabolizing genes
A folic acid metabolism gene and genotyping technology, applied in the field of molecular biology, to reduce the possibility of late contamination and improve detection efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0045] This embodiment designs folic acid metabolism genotyping primer probes, using the sequence shown in SEQ ID NO. The sequence shown in SEQ ID NO.2 at the 1298 site of the gene is used as a template to design specific primers for polymorphism detection at the 1298 site A / C, and the sequence shown in SEQ ID NO.3 at the 66 site of the MTRR gene is used as a template Design specific primers for polymorphism detection of 677 site A / G. The specific sequence is as follows:
[0046] 5'-CTCCTGACTGTCATCCCTATTG-3', the sequence is shown in SEQ ID NO.4;
[0047] 5'-AAGCTGCGTGATGATGAAATCGgCTCC-x, the sequence is shown in SEQ ID NO.5;
[0048] 5'-AAGCTGCGTGATGATGAAATCGaCTCC-x, the sequence is shown in SEQ ID NO.6;
[0049] 5'-ATGTCCACAGCATGGAGG-3', the sequence is shown in SEQ ID NO.7;
[0050] 5'-GAGGAGCTGACCAGTGAAGaAAGT-x, the sequence is shown in SEQ ID NO.8;
[0051] 5'-GAGGAGCTGACCAGTGAAGcAAGT-x, the sequence is shown in SEQ ID NO.9;
[0052] 5'-GCCTTGAAGTGATGAGGAG-3', the s...
Embodiment 2
[0061] In this example, a folic acid metabolism genotyping kit was prepared.
[0062] Since the same probe and different specific primers are used for the SNP site, the same SNP site cannot be detected in the same reaction well, otherwise it cannot be distinguished. Therefore, the present invention divides the detection of these three SNP sites into two groups, each group only detects one polymorphism of each SNP site, and the specific design of the kit is as follows:
[0063] PCR reaction premix 1: combine the primers shown in SEQ ID NO.4, SEQ ID NO.5, SEQ ID NO.7, SEQ ID NO.8, SEQ ID NO.10 and SEQ ID NO.11, wherein , the concentration of primers for SEQ ID NO.4, SEQ ID NO.7, and SEQ ID NO.10 is 0.25 μM; the concentration of primers for SEQ ID NO.5, SEQ ID NO.8, and SEQ ID NO.11 is 0.1 μM. The sequences shown in the probes SEQ ID NO.13-SEQ ID NO.15 are combined together, and the concentration of the probes is 0.1 μM. It is prepared by adding universal human-derived internal...
Embodiment 3
[0073] In this example, the folic acid metabolism genotyping kit prepared in Example 2 is used to detect samples, and the specific detection steps are as follows.
[0074] (1) Genomic DNA template extraction: Genomic DNA in blood or saliva was extracted using a universal kit.
[0075] (2) PCR amplification detection: use the genomic DNA extracted in step (1) as a template, and use the kit provided by the present invention to perform corresponding amplification detection.
[0076] Make two reaction wells for each sample, take PCR reaction master mix 1 and 21 μl of PCR reaction master mix respectively and add them to different reaction wells, then add 2 μl of the same template and 2 μl enzyme mixture, and make a negative control well and a well at the same time For positive control wells, add positive quality control product 1 to PCR reaction master mix 1 as the positive control of the reaction system, add positive quality control product 2 to PCR reaction master mix 2 as the po...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com