Method for rapid identification of Edwardsiella fisicida sRNA
A fast, suicide plasmid technology, applied in the direction of biochemical equipment and methods, microbial determination/inspection, etc., to achieve the effect of easy wide application, high-throughput detection and identification, and reduced detection cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 2
[0035] Embodiment 2 real-time fluorescent quantitative PCR analysis
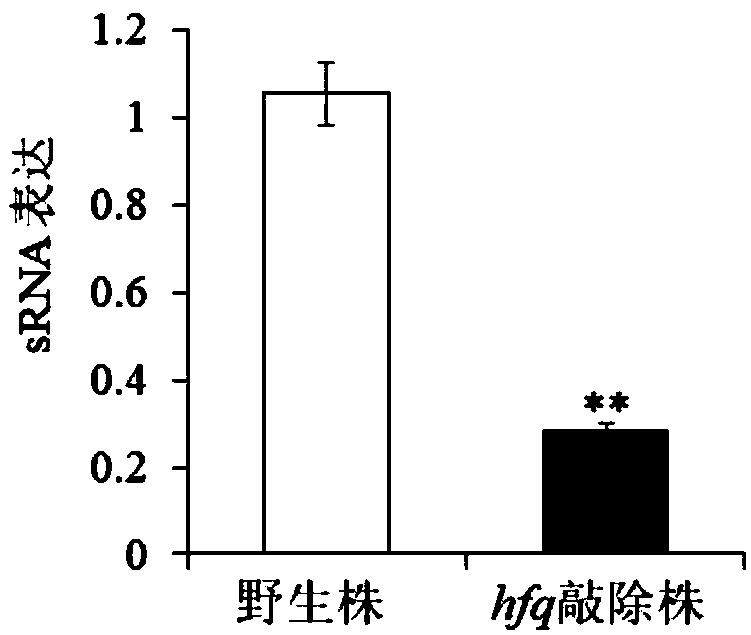
[0036] In this example, real-time fluorescent quantitative PCR was used to analyze the expression difference of sRNA082 between the wild strain and the mutant strain.
[0037] Methods: The wild strain of Edwardsiella piscicida and the hfq mutant strain were cultured to the logarithmic phase (OD=0.8), the bacteria were collected, and the RNA was extracted with the EZNA Total RNA Extraction Kit from Omega Company, and the RNA was treated with RNase-free DNaseI. Take 1 μg of RNA sample, and use Invitrogen’s Superscript II reverse transcriptase to perform reverse synthesis of cDNA; use cDNA as a template for real-time fluorescent quantitative PCR: the reagent is Takara PrimeScript TM RT-PCR KitII; reaction parameters: pre-denaturation at 95°C for 30s, denaturation at 95°C for 10s, annealing and extension at 60°C for 30s, 40 cycles; sRNA primers (taking Edwardsiella ichthyophila sRNA012 as an example), 082RTF:AA...
Embodiment 3
[0039] Embodiment 3 Accuracy detection test
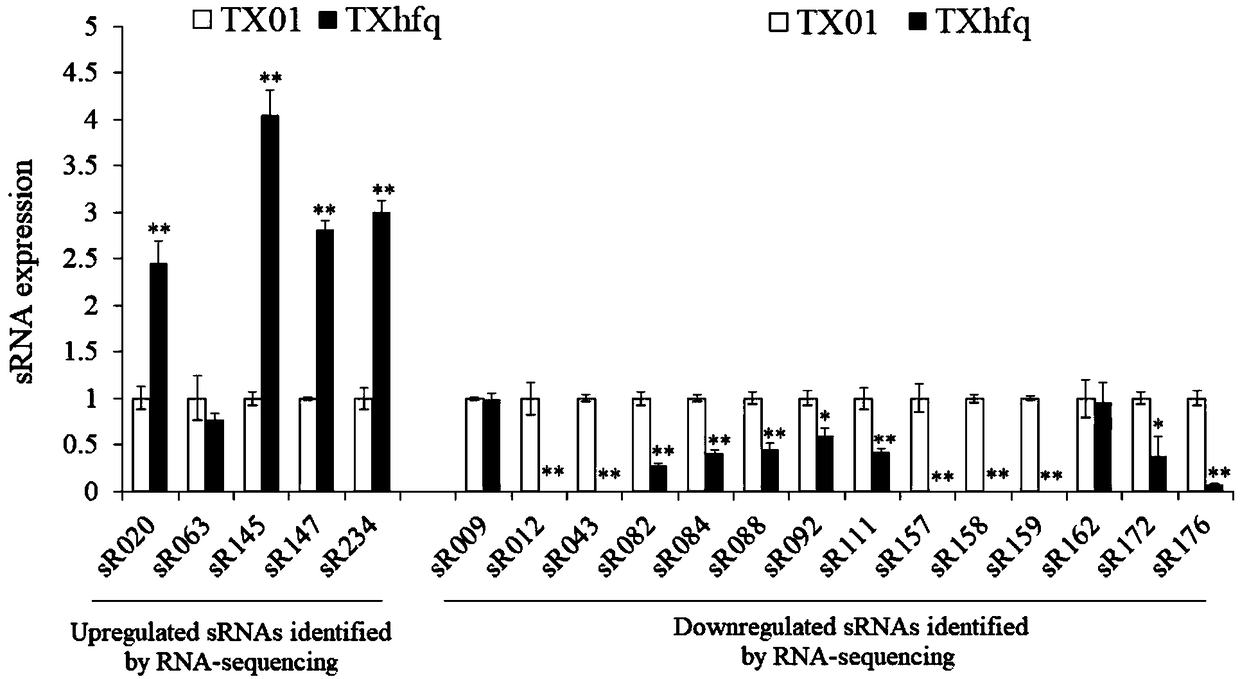
[0040] Using the method of Example 1 of the present invention and the real-time fluorescent quantitative PCR experiment conditions of Example 2, 19 sRNA samples were identified, among which, 19 samples were determined to be Hfq-dependent sRNA by transcriptome sequencing. see results figure 2 .
[0041] The results show that the method provided by the present invention detects the above 19 samples, and determines that 16 samples are Hfq-dependent sRNA, and three samples of sR063, sR009, and sR162 are Hfq-dependent sRNA, with an accuracy rate of 84.2%. It shows that the method provided by the invention can quickly and accurately identify the dependence relationship between sRNA and Hfq protein.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com