A kind of SNP molecular marker related to the background color of watermelon peel and its application
A molecular marker and watermelon technology, applied in the biological field, can solve the problems of molecular marker research reports, shorten the breeding process, and few molecular markers, and achieve the effects of improving breeding efficiency, stable amplification effect, and high accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] Example 1 Obtaining of SNP molecular marker sites
[0026] 1.1 Construction of high-density genetic map
[0027] The female parent (9904) with dark green peel background and the male parent (Handel) with green peel background were used to construct a recombinant inbred line population containing 126 individuals through continuous multi-generational selfing. The recombinant inbred line population was sequenced, high-density SNPs were developed using HighMap software, and a high-density genetic map of watermelon was constructed. A total of 178,762 SNP molecular markers suitable for recombinant inbred lines with a depth of not less than 4X were detected between the parents. Through bioinformatics analysis, it was found that the localization interval related to the background color of watermelon peel was located in the region of 2.07Mb at the end of chromosome 8 of the watermelon genome.
[0028] 1.2 Acquisition of SNP sites
[0029] CAPS markers were designed using singl...
Embodiment 2
[0030] Embodiment 2 utilizes this SNP molecular marker to identify the method for the background color of watermelon peel
[0031] 2.1. Extraction of watermelon genomic DNA
[0032] The DNA of watermelon samples to be tested was extracted by conventional CTAB method to remove RNA contamination, and the DNA sample volume was 100 μL. Determination of OD of DNA samples by UV spectrophotometer 260 / 280 Ratio, the value should be between 1.8 and 2.0 to obtain high-quality DNA samples, and the concentration should be diluted to 100ng / μL.
[0033] 2.2. Primer design
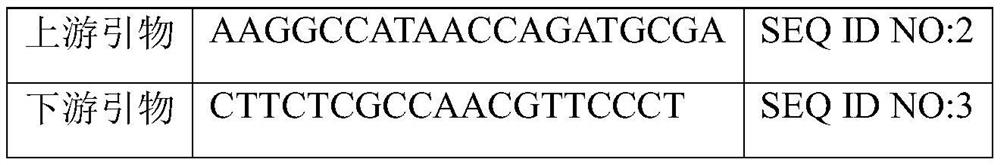
[0034] The primers were designed according to the 400bp sequence around the 29880100th base of the No. 8 chromosome of the watermelon genome. For the specific nucleotide sequence, see SEQ ID NO: 1 in the sequence table, and for the primer sequences, see Table 1.
[0035] Table 1 Primer Sequence
[0036]
[0037]
[0038] 2.3 PCR reaction system
[0039] The PCR reaction system is shown in Table 2.
[0040] T...
Embodiment 3
[0049] Example 3 Identification of 105 watermelon germplasm resources using SNP molecular markers
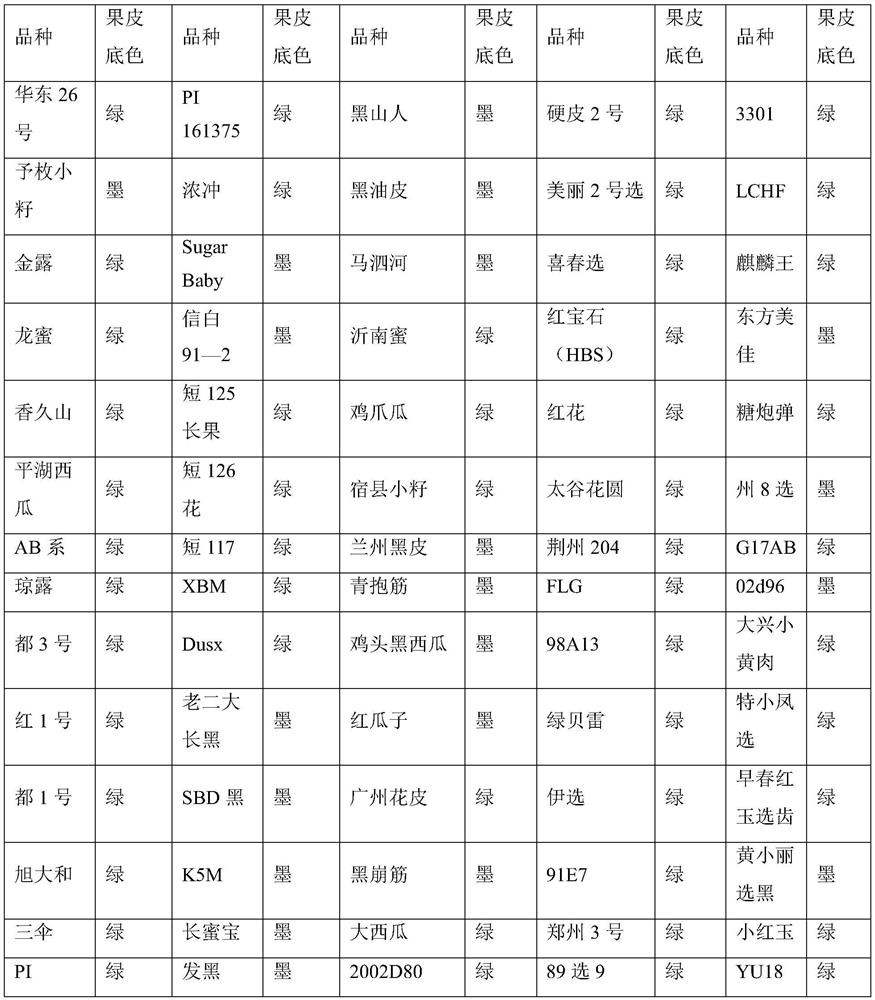
[0050]Using 105 germplasm resources of watermelon with different peel background colors in the National Watermelon Medium-term Bank as materials, the SNP molecular marker of the present invention was used to identify the peel background color. For the specific identification method, refer to the method in the above-mentioned Example 2. The identification results show that the phenotypes of the above 105 watermelon materials are completely consistent with the genotype results, indicating that the SNP molecular marker of the present invention can accurately identify the dark green color of watermelon and the background color of green peel, with an accuracy rate of 100%.
[0051] Among the 105 germplasm resources, there are 31 watermelons with dark green peel background and 74 watermelons with green peel background. The specific varieties and phenotypes are shown in Table 4 below, ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com