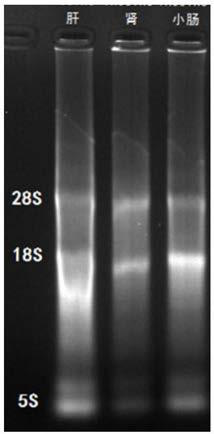
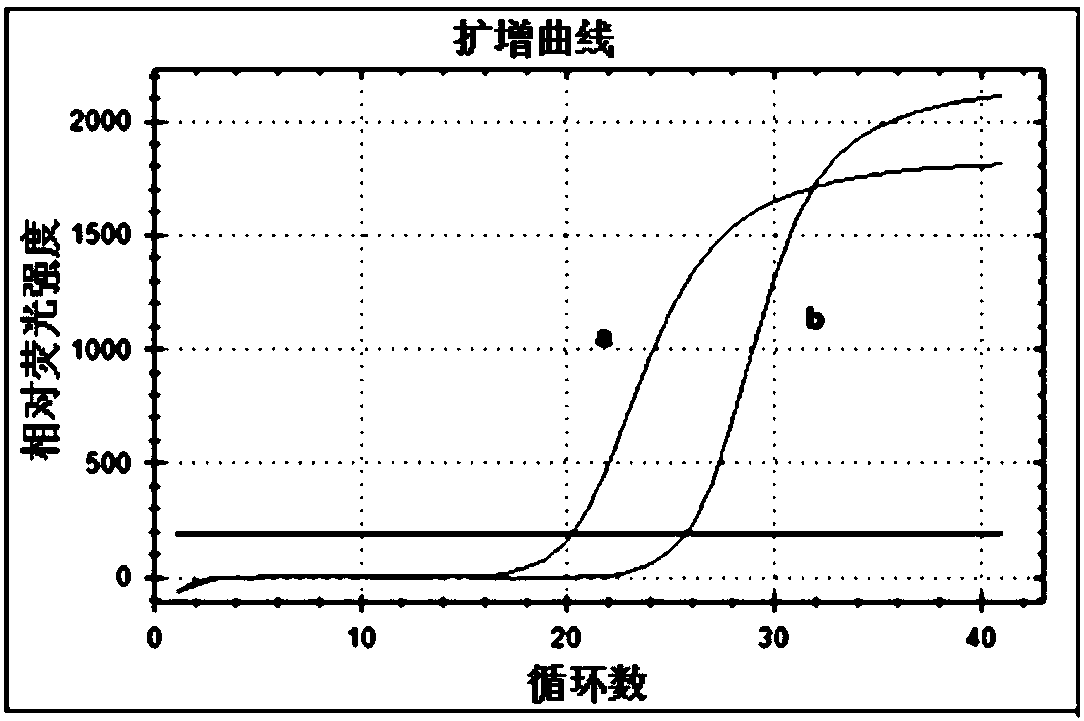
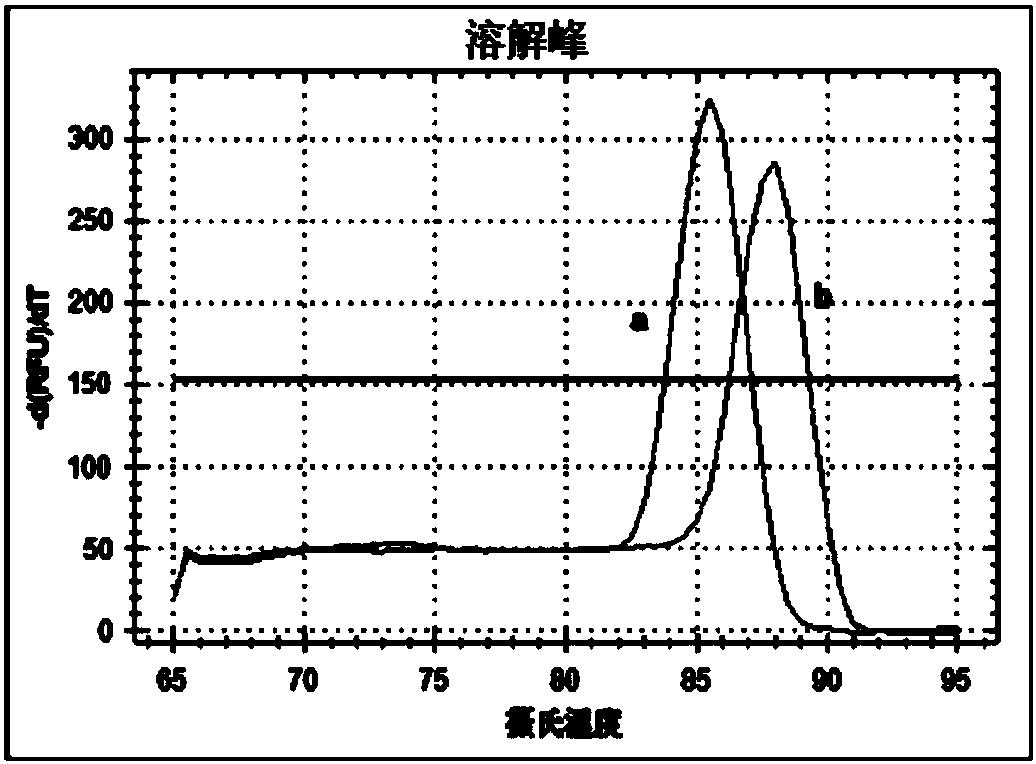
Method for detecting transcription level of SLC2A9/GLUT9 gene of tree shrews through RT-qPCR
A technology of gene transcription and gene expression level, applied in the field of detection, to achieve the effect of simple method, high sensitivity and high repeatability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0051] A RT-qPCR detection tree shrew SLC2A9 / The method of GLUT9 gene transcription level, comprises the following steps:
[0052] (1) Design the following primers:
[0053] According to the common sequence numbers of human, mouse and macaque SLC2A9 / GLUT9: human NM_020041.2, NM_001001290.1; mouse NM_001102414.1, NM_001012363.2, NM_001102415.1, NM_145559.2; macaque XM_015137992.1 primers were designed. of GAPDH Nucleotide sequence design primers were used as internal reference genes, gene accession number NM_001195426.1, primers were designed with Primer premier 5.0 software, and synthesized by Shanghai Bioengineering Co., Ltd.:
[0054] tree shrew SLC2A9 / GLUT 9 The specific upstream and downstream primers for gene expression level are:
[0055] SLC2A9 / GLUT9 F: 5'-ACAATGAAGCAGGAGCGACA-3'
[0056] SLC2A9 / GLUT9 R: 5'-ACTCAGGGTGATGTACGGGA-3'
[0057] The specific upstream and downstream primers of the tree shrew GAPDH gene as an internal reference gene are:
[0058...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com