A fluorescent biosensor for detecting dna glycosylase udg and its preparation method
A biosensor and glycosylase technology, applied in the field of biosensors, can solve the problems of long detection period, low specificity and sensitivity, and achieve the effects of fast detection speed, easy operation and short detection cycle
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
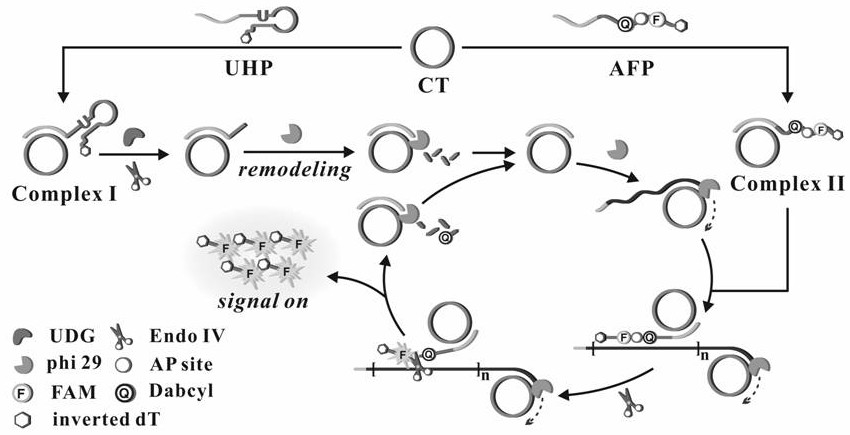
[0062] Embodiment 1 The preparation of circular template and composite probe
[0063] Prepared with 50mM Tris-HCl, 10mM MgCl 2 , T4 DNA Ligase Reaction Buffer with 10 mM DTT and 1 mM ATP. Formulated with 10mM Na 2 HPO 4 ,10mM NaH 2 PO 4 , 140mM NaCl, 1mM KCl, 1mM MgCl 2 ,1mM CaCl 2 , PBS buffer at pH=7.4.
[0064] (1) Mix 42 μL sterilized water, 6 μL linear template (10 μM), 6 μL ligation probe (10 μM) and 6 μL 10× T4 DNA ligase buffer, denature at 95°C for 5 minutes, then cool slowly to room temperature to complete hybridization , then add 3 μL T4 DNA ligase (60U / μL) to the reaction system, and react it at 16°C for 20 hours; after that, the reaction system is placed in a water bath at 65°C for 15 minutes to inactivate the T4 DNA ligation in the system enzyme.
[0065] (2) Add 3 μL of exonuclease I (20 U / μL) and 3 μL of exonuclease III (100 U / μL) to the above reaction system and react at 37 ° C for 2 h; then heat the reaction system in a water bath at 85 ° C for 10 mi...
Embodiment 2
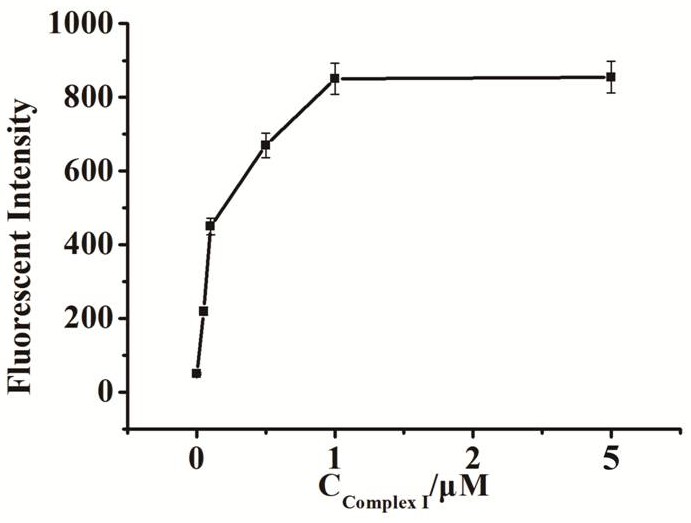
[0067] Embodiment 2 Fluorescence intensity changes with the compound probe I concentration
[0068] A method for preparing a fluorescent biosensor of the present invention, comprising the following steps:
[0069] (1) Mix 2 μL compound probe I (concentrations are 50 nM, 100 nM, 500 nM, 1 μM, 5 μM), 2 μL dNTP (1 mM), 2 μL phi29 DNA polymerase (1 U / μL), 2 μL endonuclease IV (1 U / μL) μL) in 2μL buffer (50mM Tris-HCl, 10mM MgCl 2 ,10mM (NH 4 ) 2 SO 4 , 4mM DTT, pH 7.5), add 2μL UDG enzyme solution (1U / mL) after mixing, and react at a constant temperature of 37°C for 60min after mixing;
[0070] (2) Add 2 μL of composite probe II (1 μM) to the solution in step (1), mix and react at a constant temperature of 37°C for 60 minutes;
[0071] (3) Dilute the solution obtained in step (2) with water to 100 μL, and then perform fluorescence detection; the excitation wavelength is set to 486 nm, the emission wavelength is 518 nm, and the detection range is 450 nm-530 nm, and the fluores...
Embodiment 3
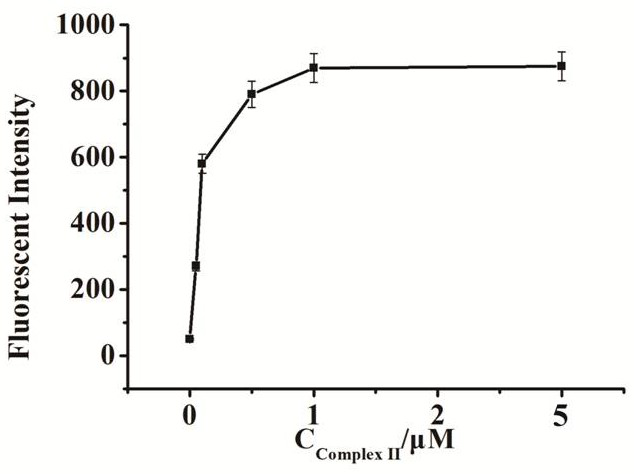
[0075] Embodiment 3 Fluorescence intensity changes with the compound probe II concentration
[0076] A method for preparing a fluorescent biosensor of the present invention, comprising the following steps:
[0077] (1) Mix 2 μL compound probe I (1 μM), 2 μL dNTP (1 mM), 2 μL phi29 DNA polymerase (1 U / μL), 2 μL endonuclease IV (1 U / μL) in 2 μL buffer (50 mM Tris-HCl ,10mM MgCl 2 ,10mM (NH 4 ) 2 SO 4 , 4mM DTT, pH 7.5), add 2μL UDG enzyme solution (1U / mL) after mixing, and react at a constant temperature of 37°C for 60min after mixing;
[0078] (2) Add 2 μL of composite probe II (concentrations are 50 nM, 100 nM, 500 nM, 1 μM, 5 μM) to the solution in step (1), mix and react at a constant temperature of 37 ° C for 60 min;
[0079] (3) Dilute the solution obtained in step (2) with water to 100 μL, and then perform fluorescence detection; the excitation wavelength is set to 486 nm, the emission wavelength is 518 nm, and the detection range is 450 nm-530 nm, and the fluorescen...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com