Application of gene editing method in Amycolatopsis orientalis
A kind of Amycolatopsis and gene technology, applied in the field of genetic engineering, can solve the problems of low efficiency and achieve the effect of simplifying genetic operation and improving integration efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
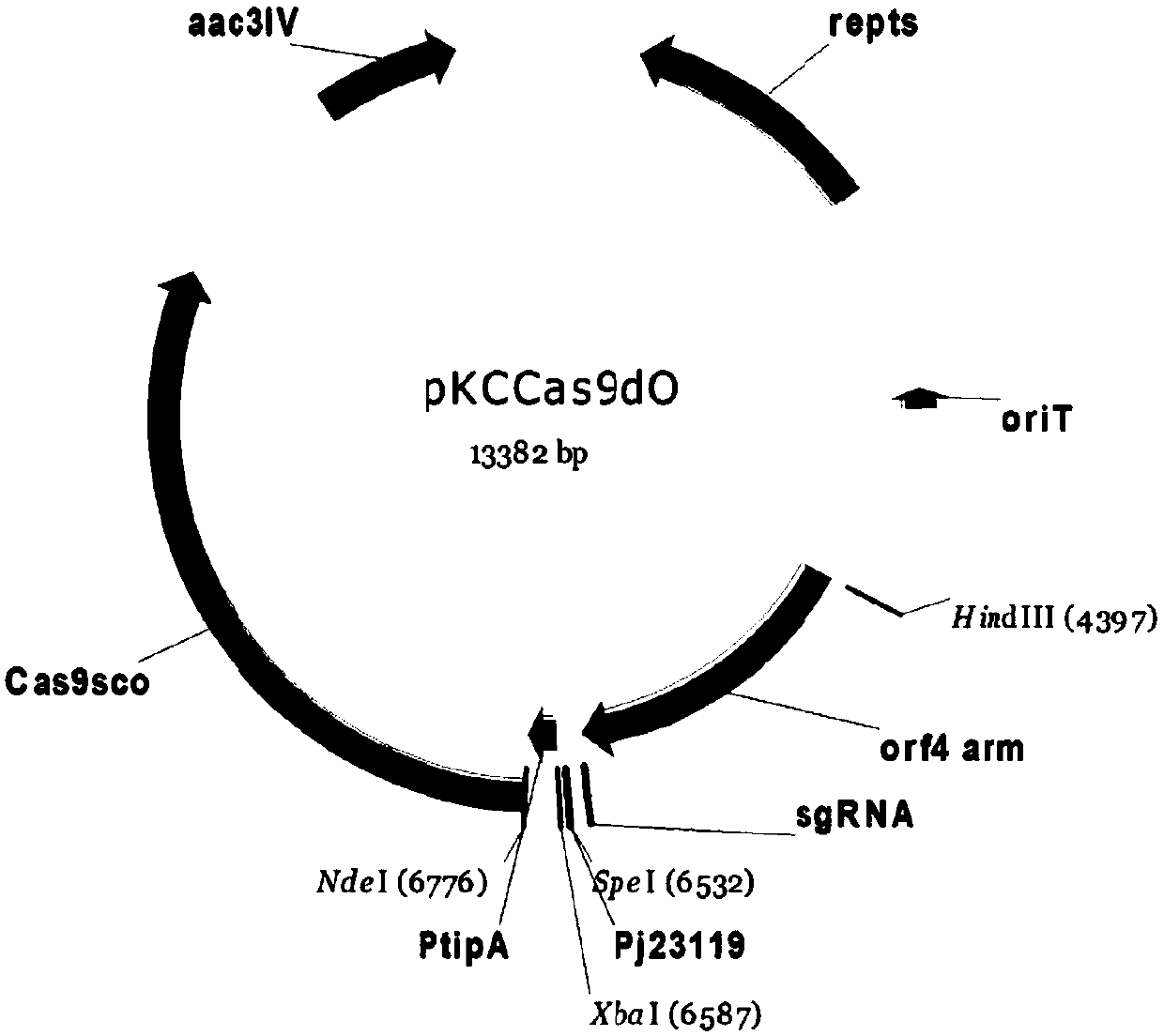
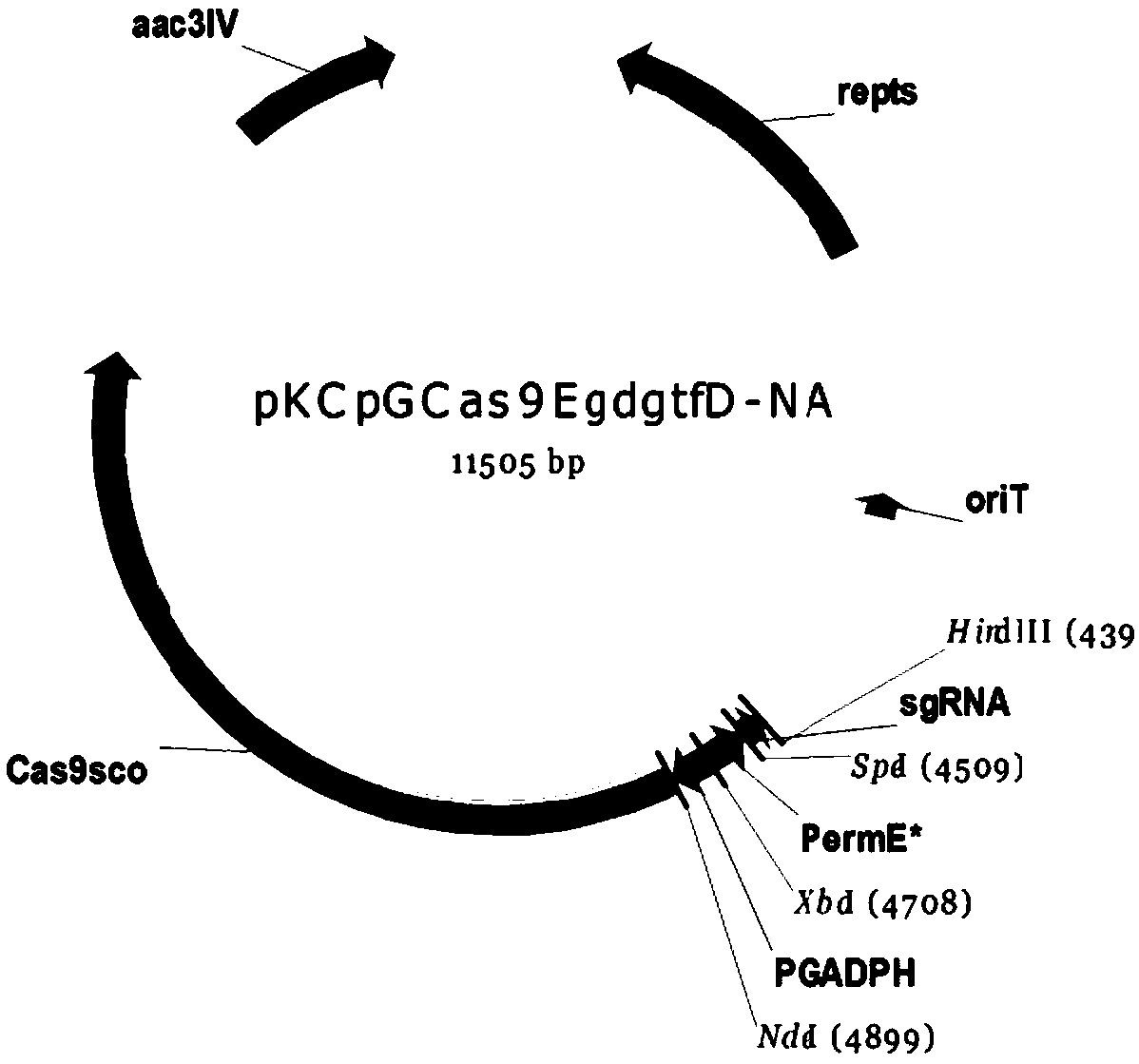
[0033] Construction and verification of embodiment 1 plasmid pKCpGcas9EgdgtfD-NA
[0034] Step 1. pKCcas9dO was digested with NdeI / HindIII to obtain the cas9sco vector. The enzyme digestion system was: 2 μL of plasmid, 0.5 μL of NdeI and HindIII endonucleases, 1 μL of Buffer, and water to 10 μL. React in a water bath at 37°C for 2 hours.
[0035] Step 2. Obtaining sgRNA recombinant fragments
[0036] The sgRNA was designed using the vancomycin-synthetic cluster glycosyltransferase gtfD gene of Amycolatopsis orientalis HCCB10007 as the editing object, and pKCcas9dO was used as the template, and the sgRNA recombination fragment was obtained by PCR amplification with primers gRNADNrecom and gtfDgRNArecom, and the fragment size was 103 bp.
[0037] gRNADN recom: ACGTTGTAAAACGACGGCCAGTGCCAAGCTTCTCAAAAAAAGCACCGAC(HindIII)
[0038] (SEQ ID NO: 1)
[0039] gtfDgRNArecom:
[0040] GGCACAATCGTGCCGGTTGGTAGGAACTAGTCGTCGAGATCGCGGTGTCGCGTTTTAGAGCTAGAAA (Spe I) (SEQ ID NO: 2)
[0041] T...
Embodiment 2
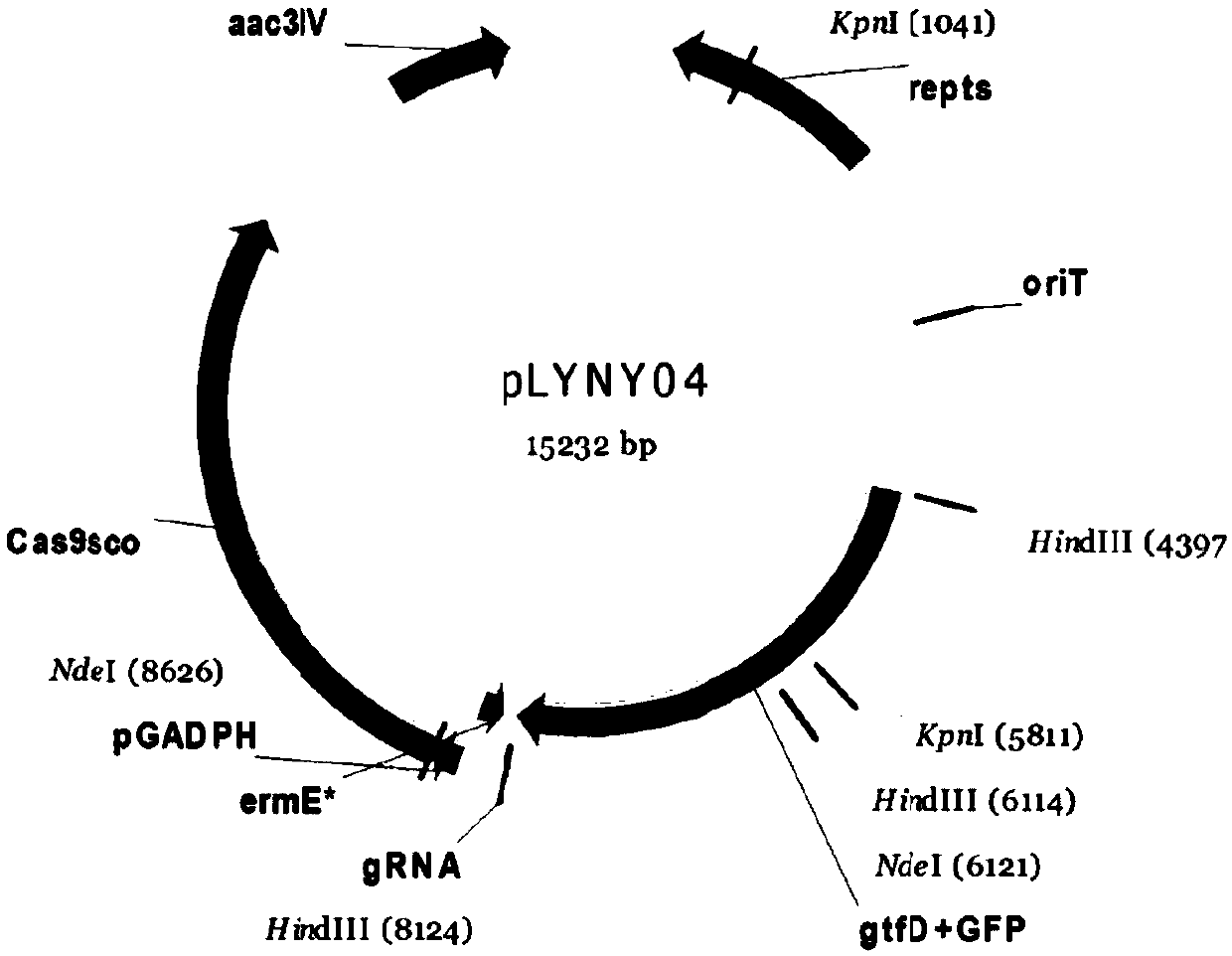
[0054] Example 2 Construction of homology arms of CRISPR / Cas9 gene editing system
[0055] Step 1, using the A.orientalis HCCB10007 genome as a template to amplify the upstream and downstream homology arm fragments with Vcm-8F / Vcm-8R and Vcm-10F / Vcm-10R respectively;
[0056] Vcm-8F: AAGCTTAGATCGGTGAGTCGCTGCTG (HindIII) (SEQ ID NO: 7)
[0057] Vcm8R: TCACGTATTTTCCCCGCTGCAGGGTACCTTCGCTACCCCTGTTTCGTG(PstI)(KpnI)
[0058] (SEQ ID NO: 8)
[0059] Vcm10F:AACAGGGGTAGCGAAGGTACCCTGCAGCGGGGAAATACGTGATGCGT(KpnI)(PstI)
[0060] (SEQ ID NO: 9)
[0061] Vcm-10R: AAGCTTTTGGTGATGATCAGGCGGGA (HindIII) (SEQ ID NO: 10)
[0062] Step 2. After the amplified upstream and downstream products are recovered by the DNA recovery kit, Overlap (overlap extension PCR method) recombination is carried out, and the obtained gtfD homology arm fragments are recovered by the DNA recovery kit, followed by T / A cloning and sequencing. The correct plasmid was recovered after digestion with KpnI / PstI;
[0063]...
Embodiment 3
[0070] Example 3 Construction of CRISPR / Cas9 editing plasmids containing gtfD gene homology arms
[0071] According to the instructions of the homologous recombination kit, the gtfD homology arm with green fluorescent protein obtained in Example 2 was integrated into pKCpGcas9EgdgtfD-NA to obtain an editing plasmid containing the homology arm of the gtfD gene. The specific operation is as follows:
[0072] Step 1. The pKCpGcas9EgdgtfD-NA backbone plasmid is linearized by HindIII digestion;
[0073] Step 2, calculating the amount of DNA required for the recombination reaction according to the formula. (To ensure the accuracy of sample addition, the sample volume of each component should not be less than 1 μl).
[0074] Linearized vector: X=[0.02*base pair number of cloning vector]ng, insert fragment: Y=Y 1 +Y 2 …+Y n =[0.04*number of base pairs per insert]ng, 2*ClonExpress Mix 5μl, add water to 10μl;
[0075] Step 3, configure the reaction system;
[0076] Step 4. Use a ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com