SNP marker co-separated from maize gray leaf spot resistant main effect QTL-qRgls1 and application of SNP marker
A gray spot disease and co-segregation technology, applied in DNA/RNA fragments, recombinant DNA technology, microbial measurement/inspection, etc., can solve the problems of unfriendly environment, high cost, low throughput, etc., to save manpower and material resources and Time, short time-consuming, and the effect of reducing detection costs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] Example 1 Acquisition of SNP markers associated with gray spot disease
[0039] It is known that the QTL locus qRgls1 related to resistance to maize gray spot disease is located on chromosome 8 of maize. The disease gene sequence is shown in SEQ ID NO: 2; Utilize the sequence alignment software DNAMAN to compare the nucleotide sequences of SEQ ID NO: 1 and 2, select 7 single nucleotide polymorphisms (SNP, Single Nucleotide Polymorphism) sites: HB1-1, HB1-2, HB1-3, HB1-4, HB1-5, HB1-6, HB1-7, and extract SNP sites HB1-1, HB1-2, HB1- 3. The nucleotide sequences of at least 50 bp on both sides of HB1-4, HB1-5, HB1-6, and HB1-7, respectively, such as SEQ ID NO: 3, SEQ ID NO: 4, SEQ ID NO: 5, SEQ ID NO: 6. Shown in SEQ ID NO: 7, SEQ ID NO: 8, and SEQ ID NO: 9. The principles for screening SNP sites are as follows: ① There are no other SNP sites in the vicinity of the SNP site; ② There are no InDel sites within 50 bp of the selected SNP site; ③ The sequences around the sele...
Embodiment 2
[0040] Embodiment 2KASP primer design and verification
[0041] The obtained SNP sites HB1-1, HB1-2, HB1-3, HB1-4, HB1-5, HB1-6, HB1-7 and the sequences on both sides such as SEQ ID NO:3, SEQ ID NO:4, SEQ ID NO: 5, SEQ ID NO: 6, SEQ ID NO: 7, SEQ ID NO: 8, SEQ ID NO: 9 (the 51st base n of each sequence is a SNP site, SEQ ID NO: 3-9 The corresponding base n is c / t, c / t, g / t, a / c, c / t, g / a, a / g (the disease resistance corresponding to each base is disease resistance / susceptibility ). Submit to Aiji Analysis (Shanghai) Technology Co., Ltd. for KASP primer design, the specific primer sequence is as follows:
[0042]
[0043] Among them, the 5' ends of Primer_FAM and Primer_HEX are respectively added with tag sequences, the tag sequence of FAM is 5'-GAAGGTGACCAAGTTCATGCT-3', and the tag sequence of HEX is 5'-GAAGGTCGGAGTCAACGGATT-3'.
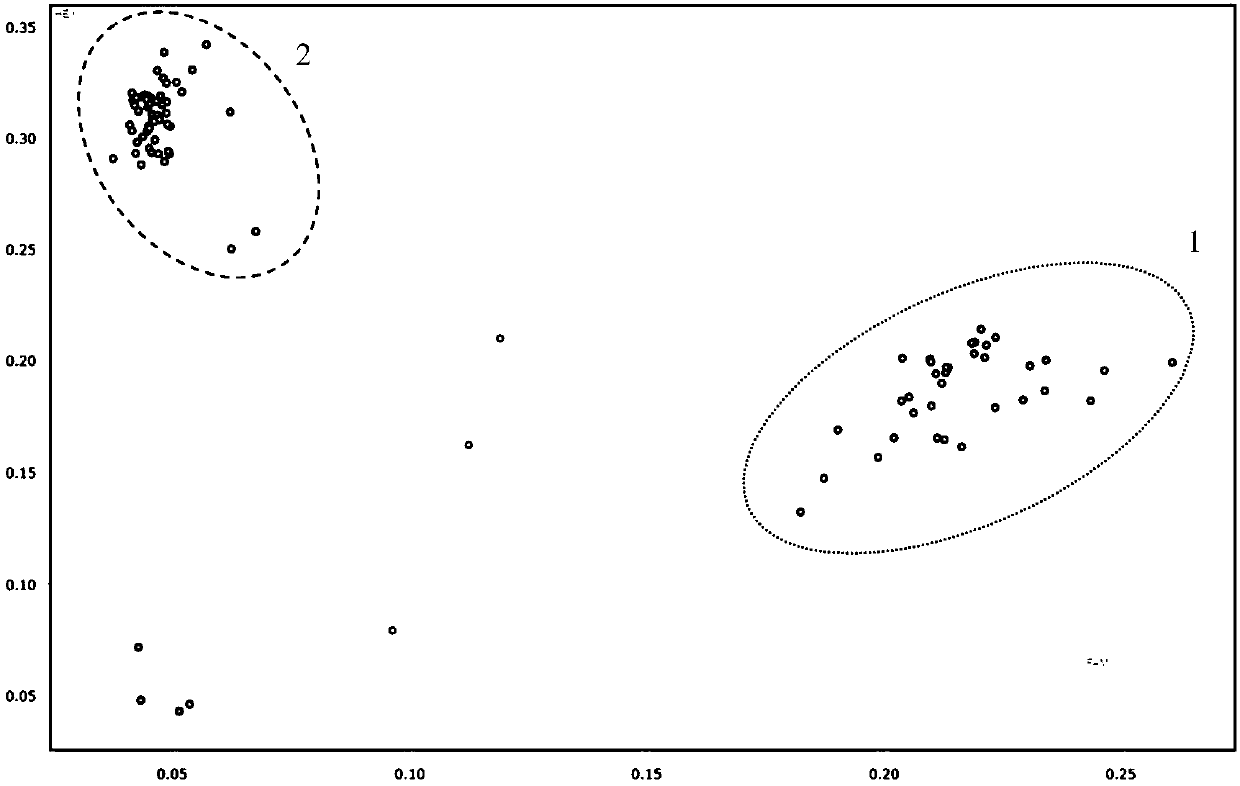
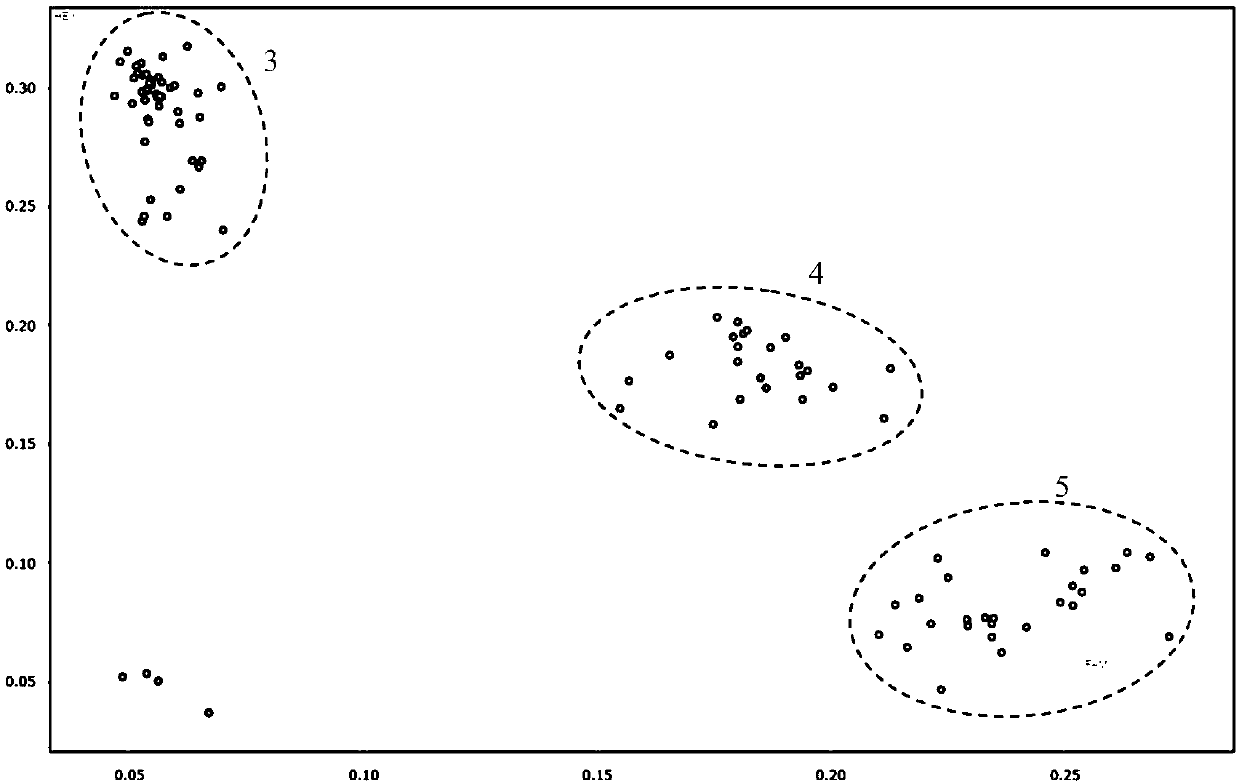
[0044] The above-mentioned KASP primers were used to detect the corresponding SNP sites, and the application effect of the KASP primers was ver...
Embodiment 3
[0045] Example 3 Use of SNP markers for assisted selection in the backcross improvement process of gray spot disease
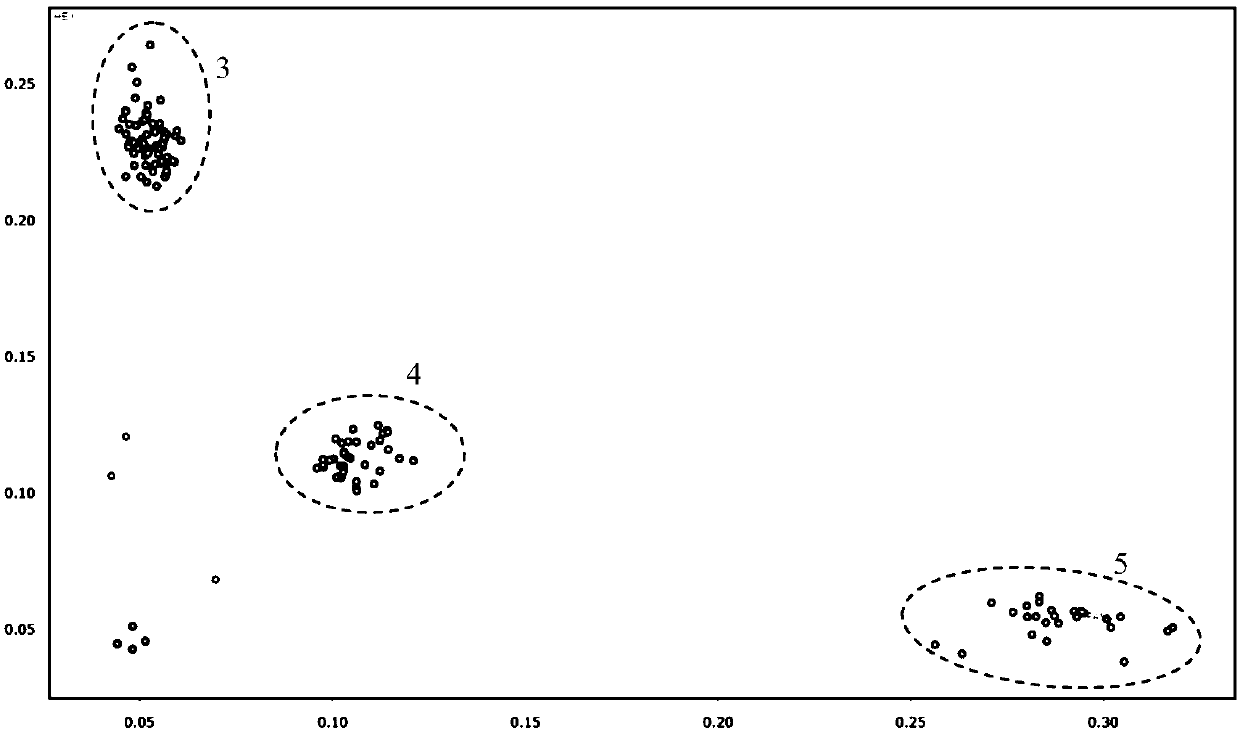
[0046] Using the disease-resistant material Y32 containing the gray spot disease resistance gene as the donor parent, and the material LA2061 susceptible to gray spot disease as the recurrent parent, the disease resistance gene was introduced into the recurrent parent LA2061 by backcrossing, and the KASP primer combination HB1- 3 and the SNP high-throughput detection platform produced by LGC for backcross generation BC 2 f 1 individual plants were tested. Specifically: take fresh leaves of each individual plant in the field, a total of 182 samples, and use the hotshot method to quickly extract leaf DNA. Use the LGCOKTOPURE DNA automatic extraction workstation to transfer the extracted DNA from the 96-well plate to the 384-well plate, use the LGCIntelliqube SNP high-throughput detection platform to divide and mix the PCR reaction system into the array tape, a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com