Kit for detecting cancer radiotherapy sensitivity and application thereof
A technology for detecting reagents and sensitivity, which is applied in the field of compositions for predicting radiotherapy sensitivity before treatment, can solve problems such as no kit detection, and achieve the effect of predicting radiotherapy tolerance/sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
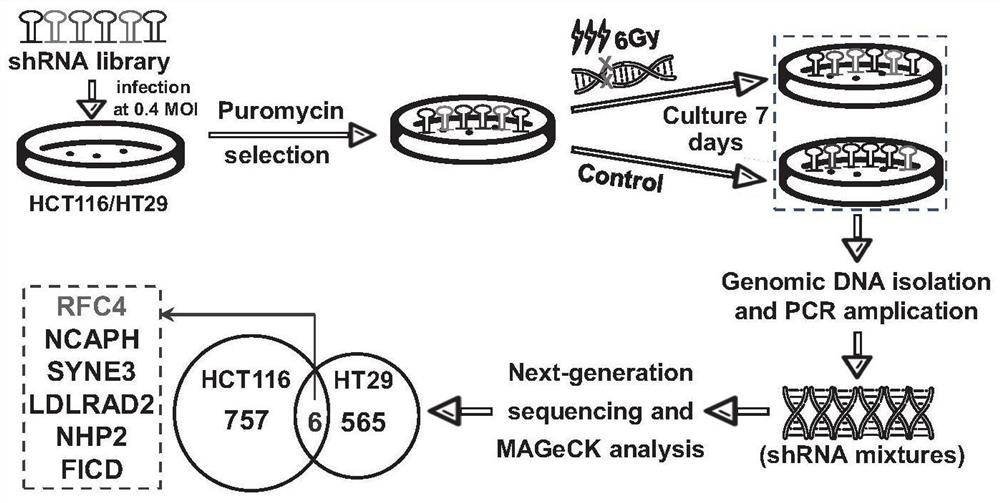
[0093] Example 1 Screening process of genome-wide shRNA library in colorectal cancer cells and construction of radiotherapy-resistant cells
[0094] The screening process is as figure 1 As shown, specifically, the entire genome shRNA library (including 75,000 shRNAs, covering about 15,000 genes) was transfected into the colorectal cancer cell line HCT116 / HT29 in the form of lentiviral vector (transfection efficiency, MOI: 0.4), After stable selection by puromycin, the cells were divided into irradiation treatment group and control group, cultured for 7 days after treatment, DNA was extracted and amplified, and the shRNA copy number change was analyzed by next-generation sequencing. Significantly down-regulated genes were selected as candidate genes.
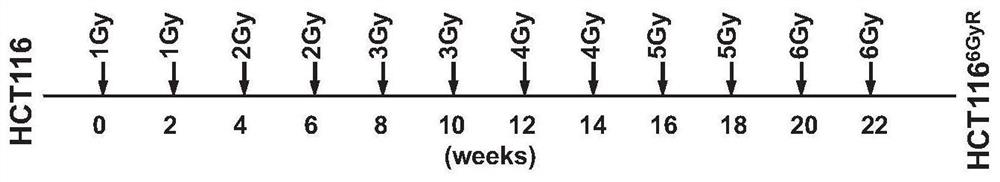
[0095] Next, if figure 2 As shown, the colorectal cancer cell line HCT116 was induced by long-term stepwise radiation to construct a stable cell line with a radiation dose of 6Gy for further screening.
Embodiment 2
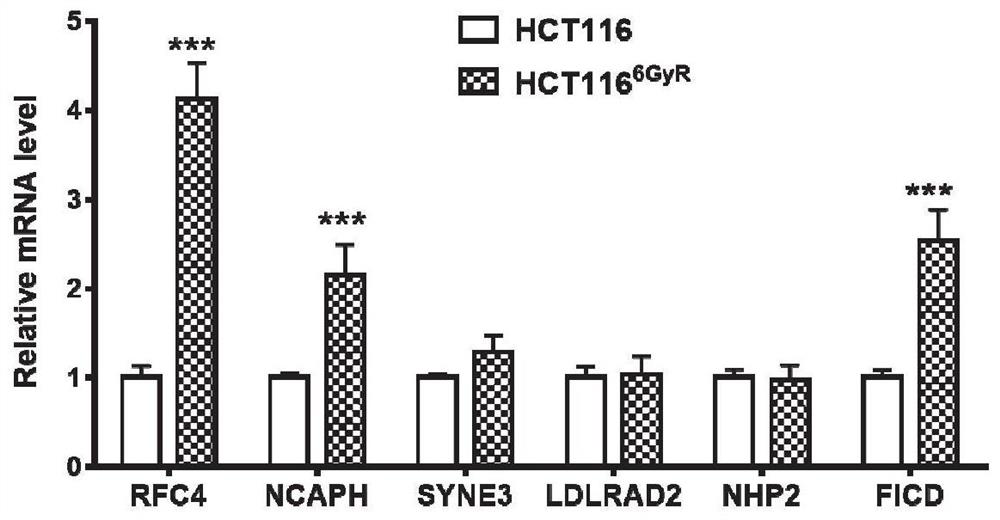
[0096]Example 2 Quantitative PCR detection of mRNA levels of candidate genes in radiation-resistant and sensitive cells
[0097] The total RNA in the cells was extracted by the Trizol method, and 2 μg of the total RNA was reverse-transcribed into cDNA. Then perform reverse transcription reaction according to the requirements of the reverse transcription system to obtain cDNA. By querying the gene database of NCBI, the cDNA sequences of RFC4 and GAPDH (internal reference) were obtained, and PCR primers were designed according to the sequences as follows:
[0098] RFC4:
[0099] The upstream primer sequence is shown in SEQ ID NO: 1,
[0100] The downstream primer sequence is shown in SEQ ID NO:2.
[0101] GAPDH (internal reference):
[0102] Upstream primer sequence: AGAAGGCTGGGGCTCATTTG,
[0103] Downstream primer sequence: AGGGGCCATCCACAGTCTTC.
[0104] Real Time PCR: According to Instructions for Premix Ex TaqTM II (TaKaRa), the formula for preparing the PCR reaction ...
Embodiment 3
[0109] Example 3 Changes in protein expression level of RFC4 before and after radiotherapy
[0110] Protein immunoassays were used to detect changes in RFC4 protein content at the cellular level. The main steps included: extraction of total protein in cells, preparation of protein loading buffer, SDS-PAGE electrophoresis, electrotransfer to PVDF membrane, and western blotting (anti-RFC4( PA5-21538) antibody (1:1000) was incubated with anti-β-actin (8H10D10, American Saixintong Company) antibody (1:1000)) and so on.
[0111] The result is as Figure 4 As shown, with the gradual increase of radiation dose, the protein level of RFC4 is also up-regulated. Combined with the results in Example 2, it shows that RFC4 is the most credible radiotherapy tolerance factor among the candidate genes.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com