Primers, kit and method for detecting SYT-SSX fusion genes
A technology of SYT-SSX1 and SYT-SSX-F, applied in the field of molecular biology, can solve the problems of poor specificity and high cost, and achieve the effects of high sensitivity, increased specificity, and strong specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0064] ARMS-QPCR kit for detecting SYT-SSX fusion gene, including qPCR reaction system, qPCR reaction system includes qPCR mixed reaction solution, reaction primers and positive control samples;
[0065] qPCR mixed reaction solution includes PCR buffer, dNTP, MgCl 2 , KOD enzyme, PCR upstream and downstream primers and TaqMan probes, primers and probes for detecting internal reference genes, and positive control samples; in the qPCR reaction system, the final concentrations of each component are: PCR buffer is 1X; dNTP is 1.5mM ; MgCl 2 2mM; KOD enzyme 0.6U / μl; PCR upstream and downstream primers 0.1μM; TaqMan probe 0.1μM; positive control sample 1ng / μl;
[0066] The sequences of PCR upstream and downstream primers and TaqMan probes are:
[0067] SYT-SSX-F: 5'-CACCACAGCCACCCCAGC-3'
[0068] SYT-SSX-TaqMan: 5'-FAM ACCAGATCATGCCCAAAGCAGCCAGC-TAMRA-3';
[0069] SSX1-R: 5'-CACTCCCTTCGAATCATTTTCG-3';
[0070] SSX2-R: 5'-CACTTCCTCCGAATCATTTCCT-3';
[0071] Primers and probes f...
Embodiment 2
[0077] Detection process:
[0078] (1) Extract tissue RNA from blood.
[0079] (2) Reverse transcription: Refer to the instructions of the Rever Tra Ace qPCR RT Kit kit from TOYOBO, and reverse-transcribe the tissue RNA in (1) into cDNA.
[0080] (3) Reagent configuration: configure fusion gene detection qPCR reaction solution and internal reference gene detection qPCR reaction solution according to the number of people to be tested;
[0081] (4) Adding samples: Take 2ul of the cDNA obtained in step (2) and add them to the fusion gene detection qPCR reaction solution and the internal reference gene detection qPCR reaction solution configured in (3); for the positive control experiment, directly add 2ul positive control substance; for negative control substance experiment, directly add 2ul negative control substance; for blank control substance experiment, add 2ul normal saline or no substance.
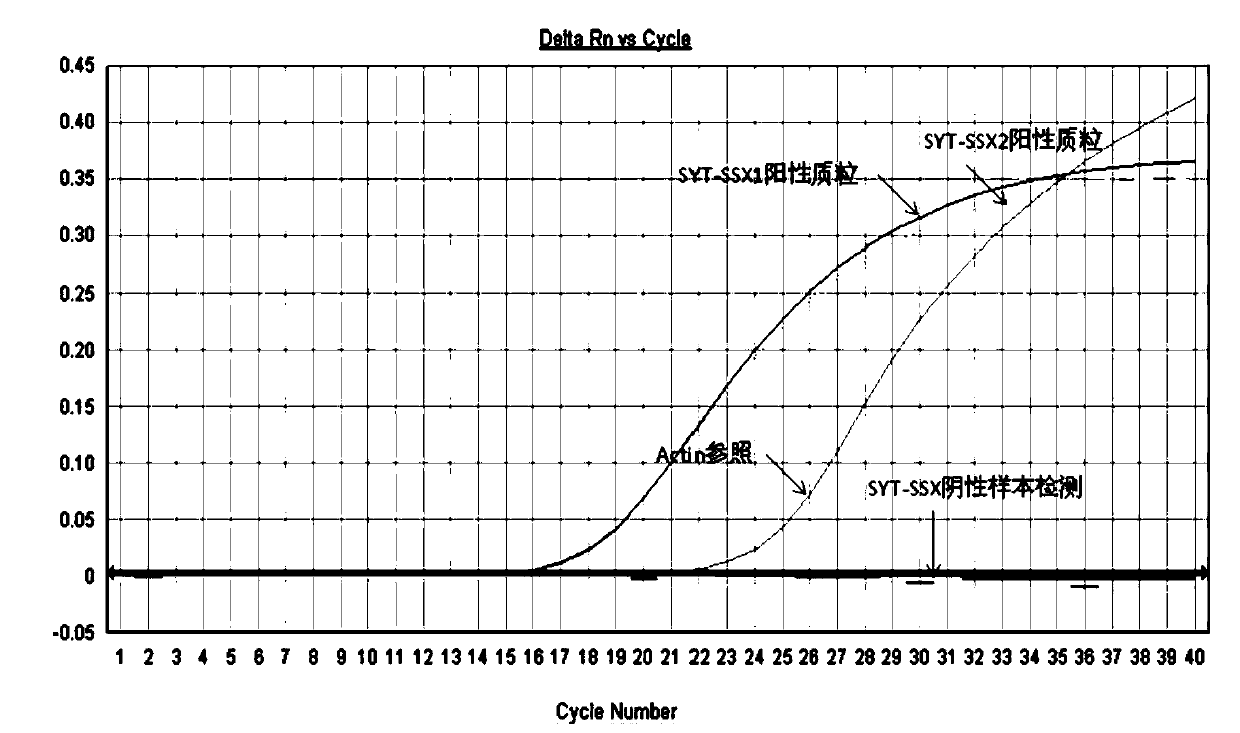
[0082] (5) Detection: detection is carried out on a real-time fluorescent PCR in...
Embodiment 3
[0089] The nucleic acid detection kit of the present invention is used to detect clinical blood samples.
[0090] Take 20 cases of clinical blood samples to be tested, extract tissue RNA in the blood samples according to the detection process described in Example 2, reverse transcribe the tissue RNA into cDNA, prepare reagents and detect.
[0091] For each sample, 2ul of the obtained cDNA was added to the SYT-SSX fusion gene detection qPCR reaction solution and the internal reference gene detection qPCR reaction solution. Carry out positive, negative and blank control experiments at the same time.
[0092] Optimization of the reaction system:
[0093] (1) Optimization of the primer concentration. Under the same conditions in the reaction system, the primer concentration was serially diluted from 0.05 μM / L to 1.5 μM / L, and the optimal concentration was determined to be 0.1 μM through the analysis of the experimental results. / L.
[0094] (2) Optimization of the probe concentra...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com