Method for protein modification
A protein modification and histone technology, applied in the field of protein modification, can solve the problems of expensive materials and cannot meet the needs of protein synthesis, and achieve the effect of shortening the reaction time and reducing the cost.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] Example 1. Thioacetylation modification of the No. 16 lysine site on H4
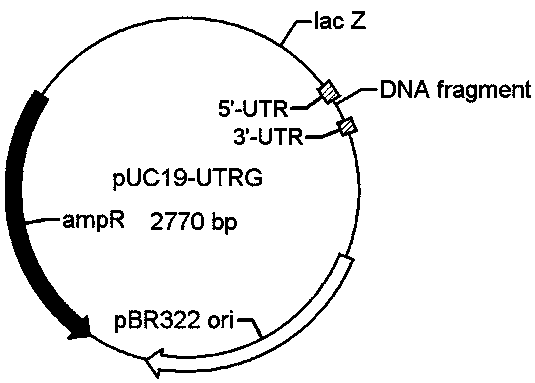
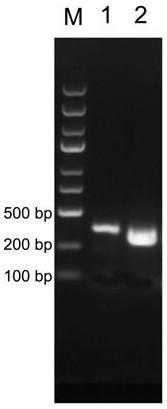
[0037] With pUC19-UTRG vector containing human histone H4 sequence (such as figure 1 Shown is the pUC19 vector map used to insert the target protein fragment. The DNA fragment site is used to insert any protein fragment) to prepare a DNA template with a lysine codon mutation site, and mutate the codon encoding lysine at position 16 of human histone H4 to an amber terminator (TAG ), as shown in SEQ ID NO:2 (source sequence) and SEQ ID NO:4 (mutated sequence). The specific method is to use the pUC19-UTRG vector containing the H4 fragment as a template to design reverse primers for mutation, and perform PCR amplification. The specific operation refers to Mutan BESTKit (TaKaRa, R401), and the amplified fragments are subjected to agarose gel electrophoresis. (Such as figure 2 As shown, the band marked 2 is the mutant fragment of H4), and purified by gel cutting, the purified fragment was end-ligate...
Embodiment 2
[0040] Example 2. Acetylation modification of lysine 27 on H3.
[0041] Use the pUC19-UTRG vector containing the human histone H3 sequence to prepare a DNA template with a lysine codon mutation site, and mutate the codon encoding lysine at position 27 of human histone H3 to an ocher terminator (TGA), as shown in SEQ ID NO: 1 (source sequence) and SEQ ID NO: 3 (mutated sequence). The specific method is to design a reverse primer for mutation using the pUC19-UTRG vector containing the H3 fragment as a template, and perform PCR amplification. The specific operation refers to the Mutan BEST Kit (TaKaRa, R401), and the amplified fragment is subjected to agarose gel electrophoresis (eg figure 2 As shown, 1 is the amplification product of the H3-K27 template), and purified by gel cutting, and the purified fragments were end-ligated with topoisomerase, and then transferred into E. coli DH5α to obtain mutants. The mutant vector was linearized and concentrated to obtain a purified pr...
Embodiment 3
[0044] Example 3. Simultaneous synthesis of H3-AcK27 and H4-ThioAcK16 in an in vitro expression system.
[0045] The mutants of Example 1 and Example 2 were used as templates.
[0046] Prepare tRNA sep and Flexizyme. For specific operations, refer to Embodiment 1.
[0047] Mix 1μl tRNA (250mM), 1μl Flexizyme (250mM), and 1μl HEPS-KOH (500mM), heat at 95°C for 3 minutes and then cool at room temperature, add 2μl ThioAcK compound (25mM), mix well and ice-bath for 6 hours to get ThioAcK-tRNA, the results were detected by Acid-PAGE.
[0048] Mix 1μl tRNA (250mM), 1μl Flexizyme (250mM), and 1μl HEPS-KOH (500mM), heat at 95°C for 3 minutes and then cool at room temperature, add 2μl AcK compound (25mM), mix well and ice-bath for 6 hours to get ThioAcK-tRNA, the results were detected by Acid-PAGE.
[0049] Using the PURE EXPRESS SYSTEM (NEB, E6800S) in vitro protein expression system, add 1 μL of H3-K27 and H4-K16 templates and 0.5 μL of ThioAcK-tRNA and AcK-tRNA synthesized in t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com