Genome targeted modification method
A targeted modification and genome technology, applied in the field of genome modification, can solve the problems of inability to construct conditional knockout at one time, and inability to achieve conditional knockout at the same time.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0199] Example 1 Creation of tbx5a conditional knockout and gene / cell marker fish lines by NHEJ-based gene knock-in
[0200] 1. Construction of tbx5a gene knock-in vector
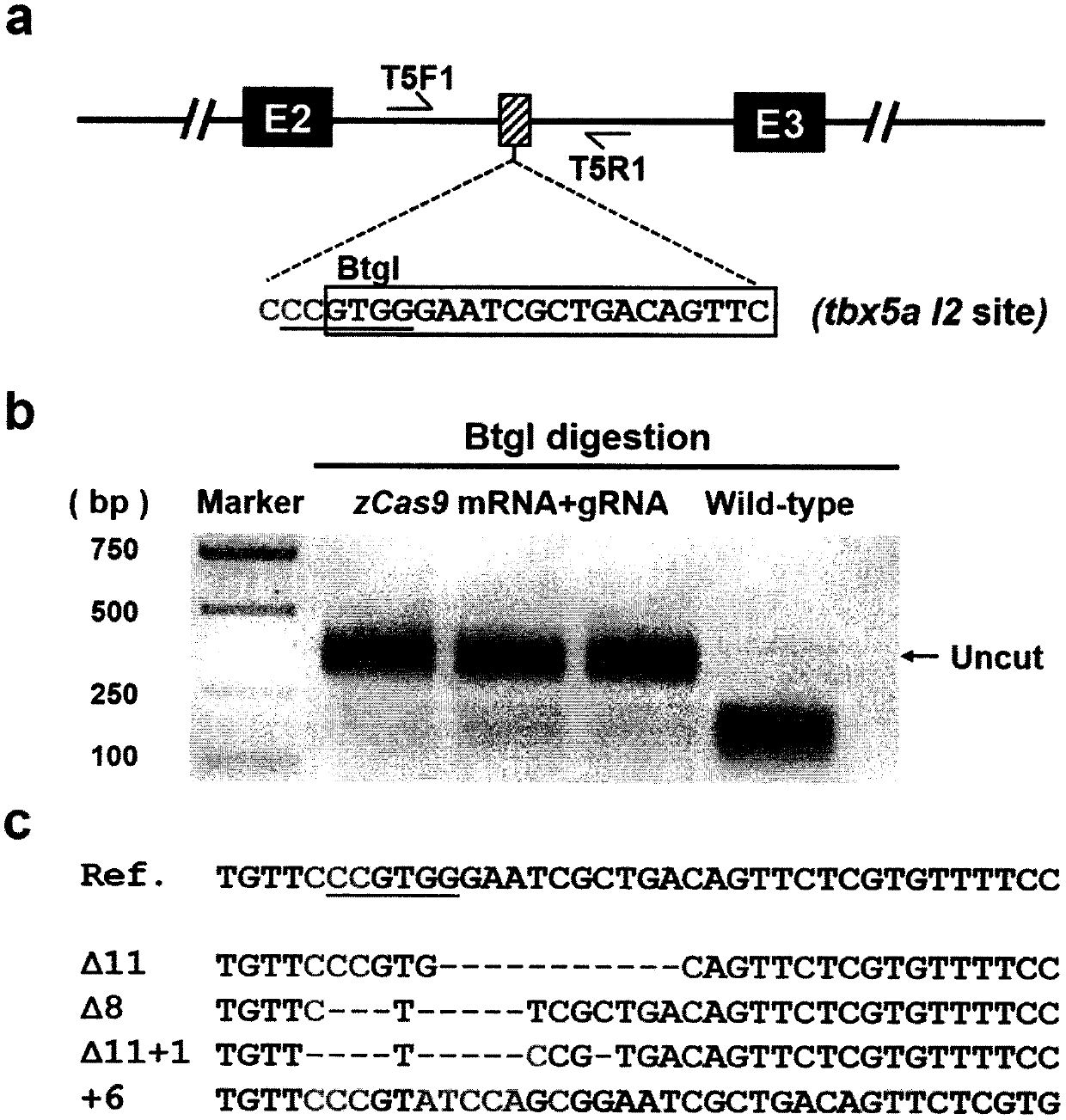
[0201] Tbx5a is an important regulatory gene of zebrafish heart development. Based on the nucleic acid database (Nucleotide, https: / / www.ncbi.nlm.nih.gov / nucleotide / ) in the NCBI website and the zebrafish genome database in Ensembl (version Zv9, http: / / asia.ensembl.org / Danio_rerio / Info / Index) analyzed the gene structure and sequence of tbx5a. There are four transcripts of tbx5a, among which the transcripts tbx5a-001 and tbx5a-202 encode the same protein with 492aa of amino acid residues. The transcript tbx5a-201 encodes a protein of 485 aa, but this transcript was removed from the NCBI database. Transcript tbx5a-002 is a non-coding RNA. In this experiment, we chose the transcript tbx5a-202 as the main reference sequence, and its sequence is shown in SEQ ID NO:5. CRISPR / Cas target design and efficiency...
Embodiment 2
[0225] Example 2 Creation of kctd10 conditional knockout and gene / cell marker fish lines by NHEJ-based gene knock-in
[0226] A CRISPR / Cas target site was designed in the first intron of kctd10, named kctd10 I1, and its sequence is shown in SEQ ID NO:10. Efficiency was evaluated using a single restriction endonuclease Hpy188I corresponding to the target, and the results showed that the kctd10 I1 site had a high cutting activity with an efficiency close to 97%, and different forms could be detected in the target sequence by sequencing the indel. Therefore, the kctd10 I1 target was selected as the gene knock-in site for subsequent experiments ( Figure 11 ).
[0227] Based on a strategy similar to Example 1, the positive-carrier kctd10-T2A-tdGFP floxP 2PA-mutExon PoNe donor( Figure 12, hereinafter referred to as kctd10 PoNe donor), its sequence is shown in SEQ ID NO:2. Then, the kctd10 gene knock-in system was injected into single-cell stage zebrafish embryos by microinject...
Embodiment 3
[0229] Embodiment 3 prepares the genotype marker fish line of tbx5a
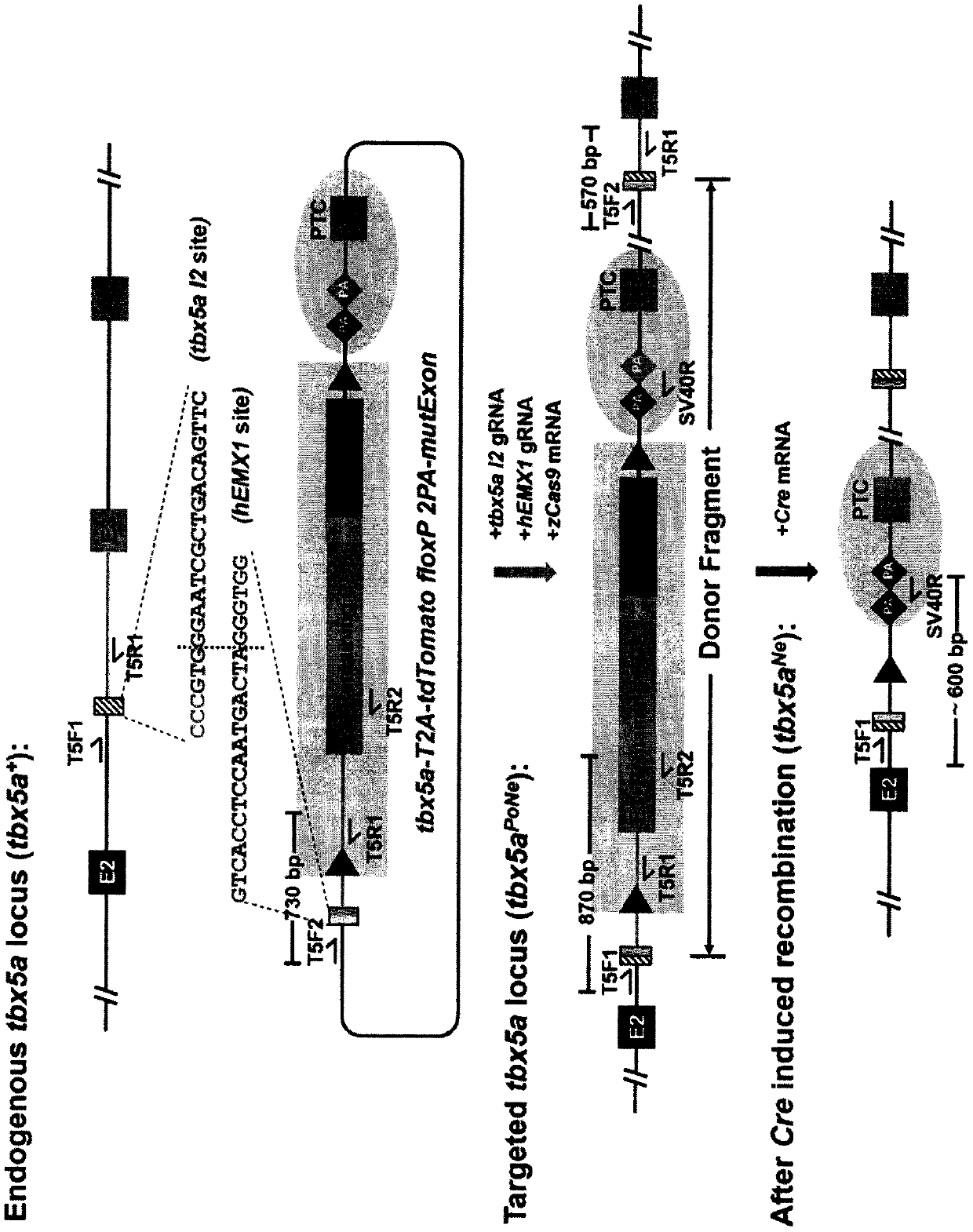
[0230] First, the tbx5a I2 target in Example 1 is also used as the gene knock-in site ( figure 1 ), constructed the genotype tagging vector of tbx5a tbx5a-T2A-tdT-2PA floxP tdG-2PA geno-tagging PoNe Donor( Figure 14 , hereinafter referred to as tbx5a geno-tagging donor), its sequence is shown in SEQ ID NO:3.
[0231] Inject gene knock-in system into single-cell stage zebrafish embryos by microinjection: zCas9 mRNA 300-400ng / μL, tbx5a I2 gRNA 100ng / μL, hEMX1 gRNA 100ng / μL, tbx5a geno-tagging donor(hEMX1) 15ng / μL , and obtained the F 0 Zebrafish embryos.
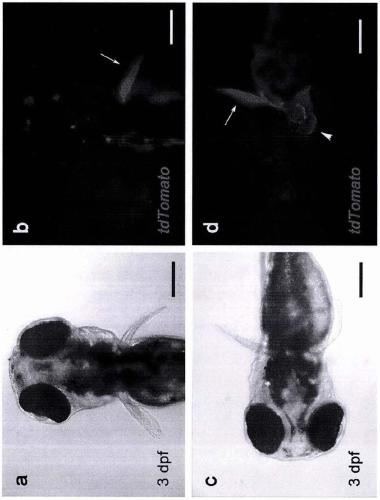
[0232] for the F 0 stage to verify the effectiveness of genotype markers as soon as possible. In another similar experiment, we also added Cre mRNA to the above gene knock-in system and co-injected zebrafish single-cell stage embryos. At this point, if only gene knock-in events have occurred, tdTomato in the positive element (Po-cassette) will produce ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com