Method for quickly detecting drug resistance of bacteria
A technology for drug resistance and bacteria, applied in biochemical equipment and methods, microbial measurement/testing, Raman scattering, etc., can solve problems such as inaccurate judgment of drug sensitivity results, and achieve accurate sensitivity effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
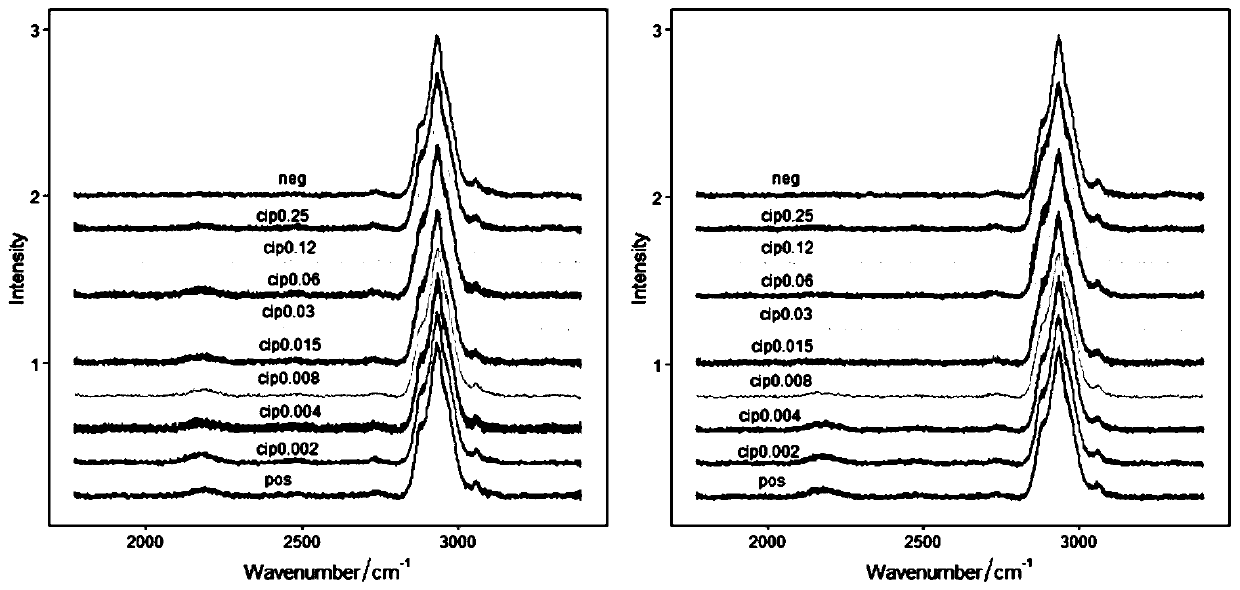
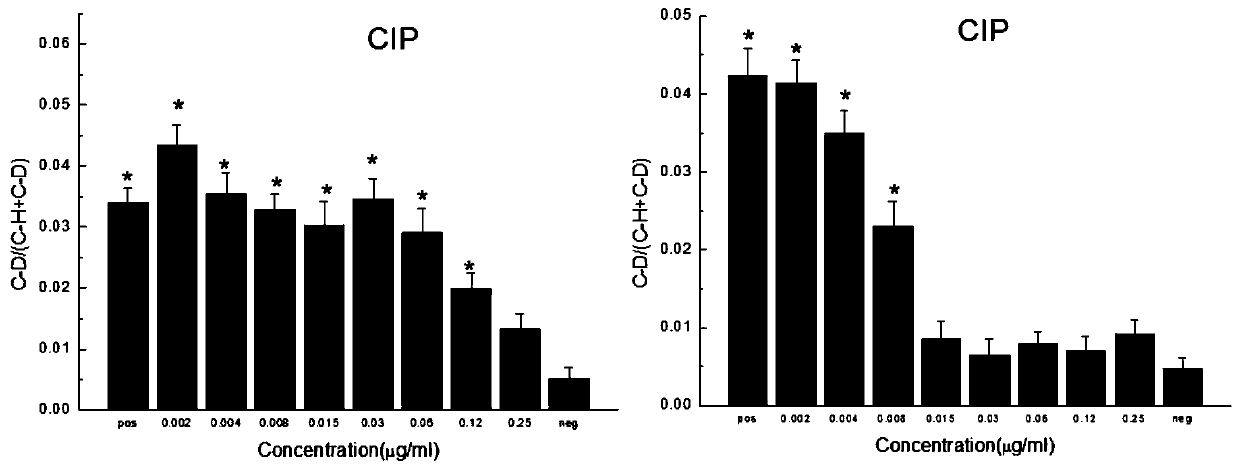
[0034] In this embodiment, Escherichia coli ATCC25922 is selected. Since its MIC quality control range is much smaller than the sensitive break point, the concentration setting only crosses its MIC quality control range.
[0035] The method for rapid detection of Escherichia coli ATCC25922 to Ciprofloxacin (Ciprofloxacin) resistance includes the following steps:
[0036] Set up different concentrations of antibiotic experimental group and positive control group and negative control group. Among them, the different concentration of antibiotic experimental group refers to adding different concentrations of antibiotics to the medium, and the positive control group refers to adding antibiotics to the medium at a concentration of 0 , The negative control group means that the concentration of antibiotics added to the medium is 0,
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com