Proteins and peptide tags with enhanced rate of spontaneous isopeptide bond formation and uses thereof
An isopeptide bond and identity technology, applied in the field of two-part joints, can solve the problems of limited reaction rate and suboptimal reaction rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0273] Example 1 - Spy Tag (SEQ ID NO:6) Phage display optimization
[0274] Spy tags / spy captures are an unconventional approach to peptide interactions, and some features of the interactions cannot be predicted by rational design. Selection from phage libraries has been around for decades, and the difficulty has generally been the detection of weak interactions rather than the challenge of screening for irreversible interactions. We initially established model selection to study efficient isopeptide bond formation selection.
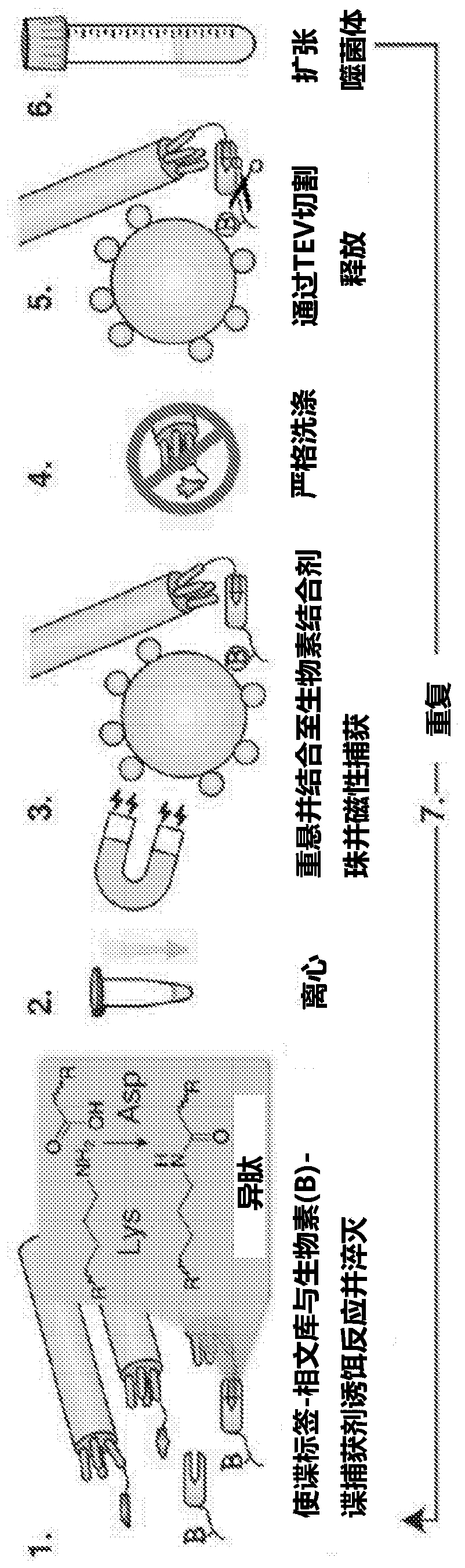
[0275] We found that the first key function to be able to successfully pan the spy tag - phage is to capture the spy capture agent (SEQ ID NO: 7) bait in the capture solution, rather than attach the spy capture agent to the beads. Solution capture reagents allow for easy titration of titrant concentrations and reduce the background of non-specific binding of phage to beads ( figure 1 ).
[0276] The second key feature is the specific elution o...
Embodiment 2
[0280] Embodiment 2-spy capture agent (SEQ ID NO:7) Phage display optimization
[0281] Phage display selection of spy capture agents is performed in a manner similar to selection of spy tag variants, although displaying split proteins on the phage surface provides a further challenge. The key features we found to be important for efficient selection are the TEV protease cleavage site between the spy capture agent and pIII on the phage (allowing specific elution of the phage from the beads) and co-translational translocation using the DsbA signal sequence, thereby Improved display of Spy Capture Agents on pIII. The bait was biotinylated Avi-tag-Spy-MBP, and the spy-capture variants were prepared by error-prone PCR ( Figure 4 A). We initially optimized model selection using the desired bait (spy tag) or negative control spy tag DA, which non-covalently binds to the spy capture agent but does not react. Recovered phage were evaluated by qPCR. This selection showed tha...
Embodiment 3
[0285] Embodiment 3-Verification of mutation rate of spy label 002 and spy capture agent 002
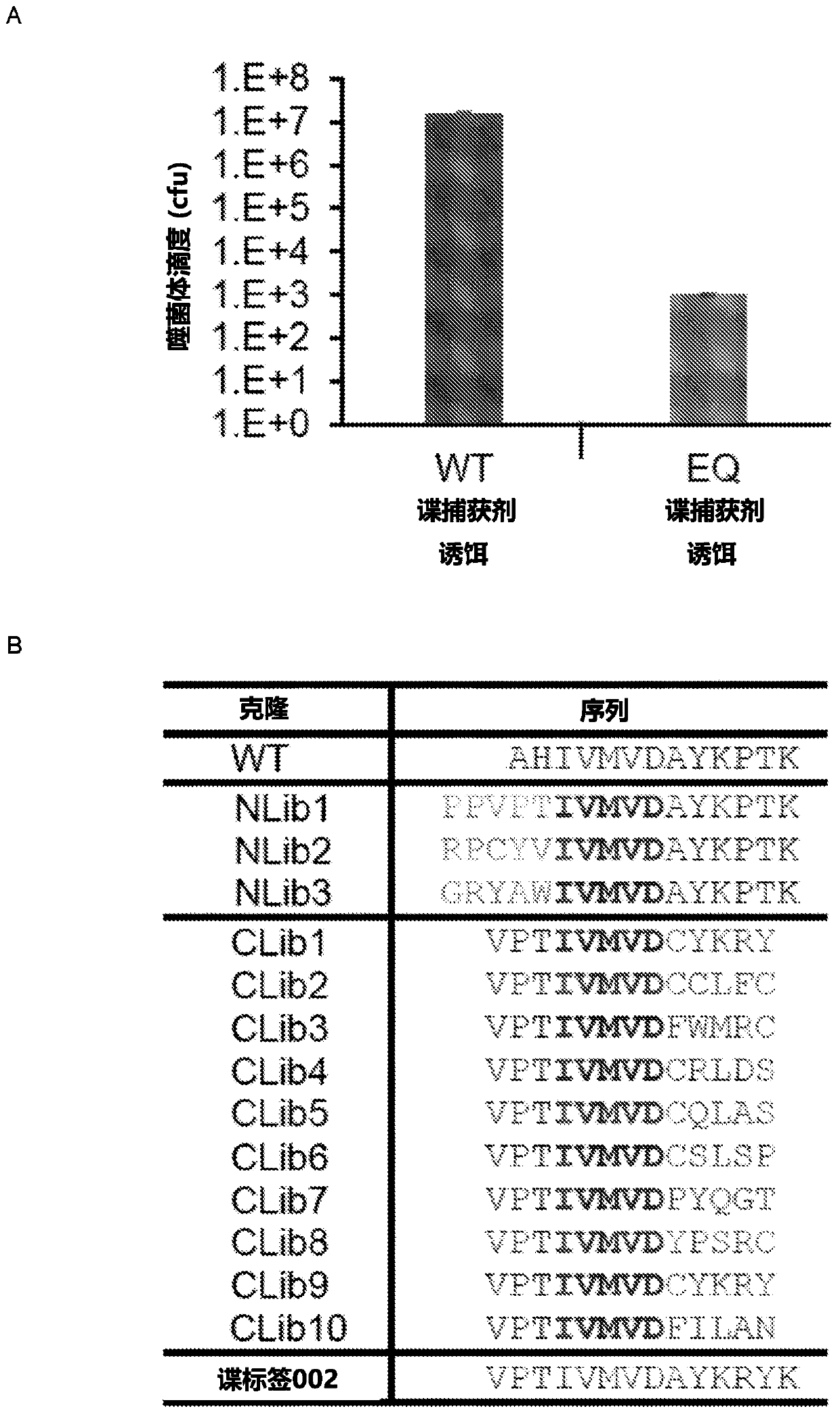
[0286] After having Spy Tag 002 and Spy Capture Agent 002, we carefully verified their response behavior. We confirmed the putative key role of reactive residues by showing the response to ablation of a single mutation in spy tag 002 (DA) or spy capture agent 002 (EQ) ( Figure 9 A).
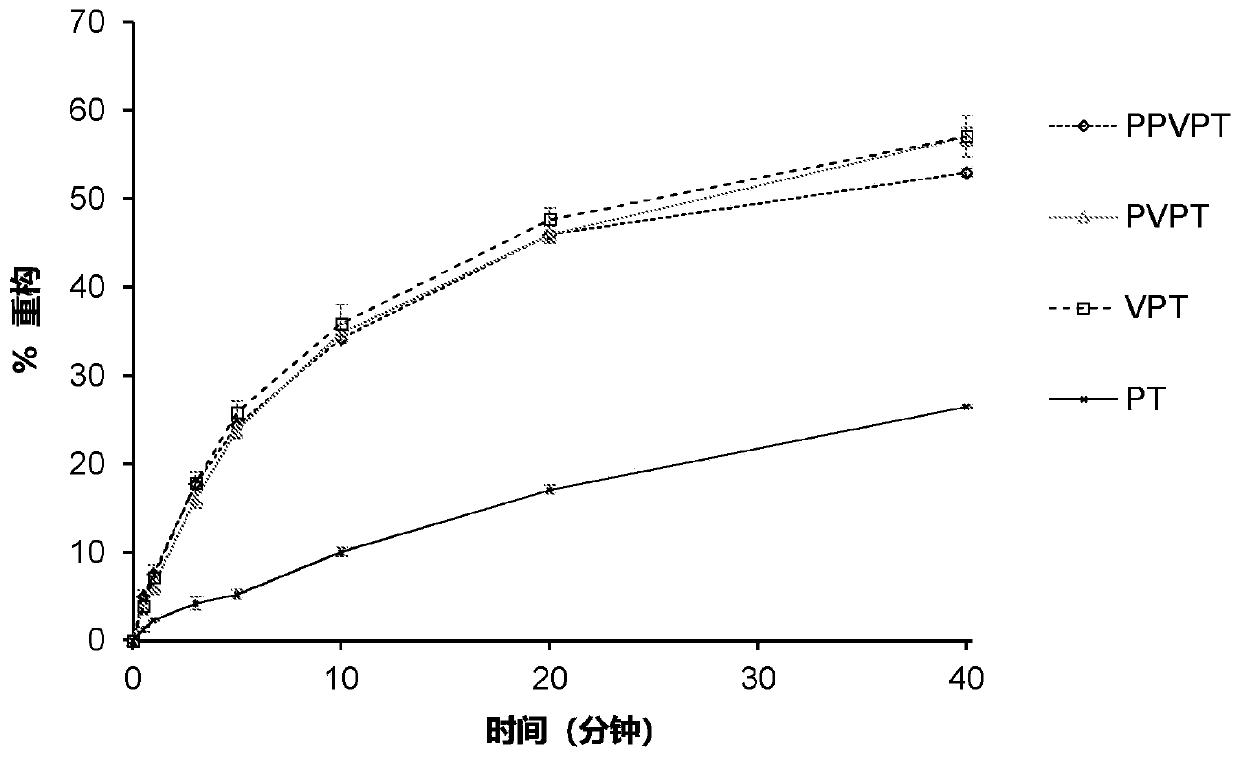
[0287] The spy tag / spy capture agent reaction is effective at high concentrations. In order to analyze the reaction at low concentrations, after polyacrylamide electrophoresis, it was reacted with superfolded GFP (superfolder GFP, sfGFP) for fluorescence detection of the formation of covalent bonds. If the sample is not boiled, sfGFP can remain folded and fluoresce even in the presence of SDS. This analysis showed that the reaction rates of SpyTag 002 and Spy Capture Agent 002 were significantly increased compared to the parental version ( Figure 9 B). As expected, the difference was smaller wh...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com