Method for identifying functional kinase for regulating cell autophagy
A kinase and cell technology, applied in the field of bioinformatics, can solve the problem of low accuracy of protein kinases, achieve the effect of narrowing the screening range, reducing workload, and accurately identifying
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
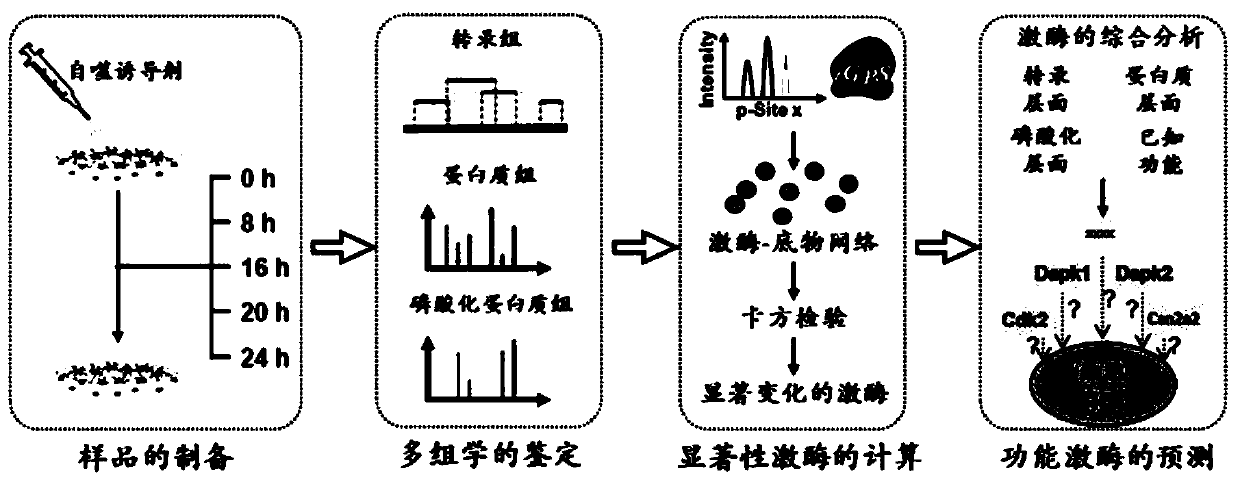
[0043] This example provides a method for predicting regulatory autophagy kinases based on multi-omics integration, such as figure 1 and figure 2 shown, including the following steps:
[0044] Using silica nanomaterials with a size of 16nm as an autophagy inducer at a final concentration of 60ug / mL, normal rat kidney cells were treated for 0 hours, 8 hours, 16 hours, 20 hours, and 24 hours to treat 0 The normal rat kidney cells for 4 hours were used as the control group, and cell samples at 4 time points after treatment were collected.
[0045] Using the second-generation gene sequencer HiSeq 4000system, RNA sequencing was performed on cell samples before and after treatment with 16nm silica nanomaterials to obtain transcriptome data.
[0046] The Bowtie-Tophat-Cufflinks series software was used to analyze and process the transcriptome data to obtain quantitative information of genes. Among them, a total of 6059 genes were significantly changed after nanomaterial treatment...
Embodiment 2
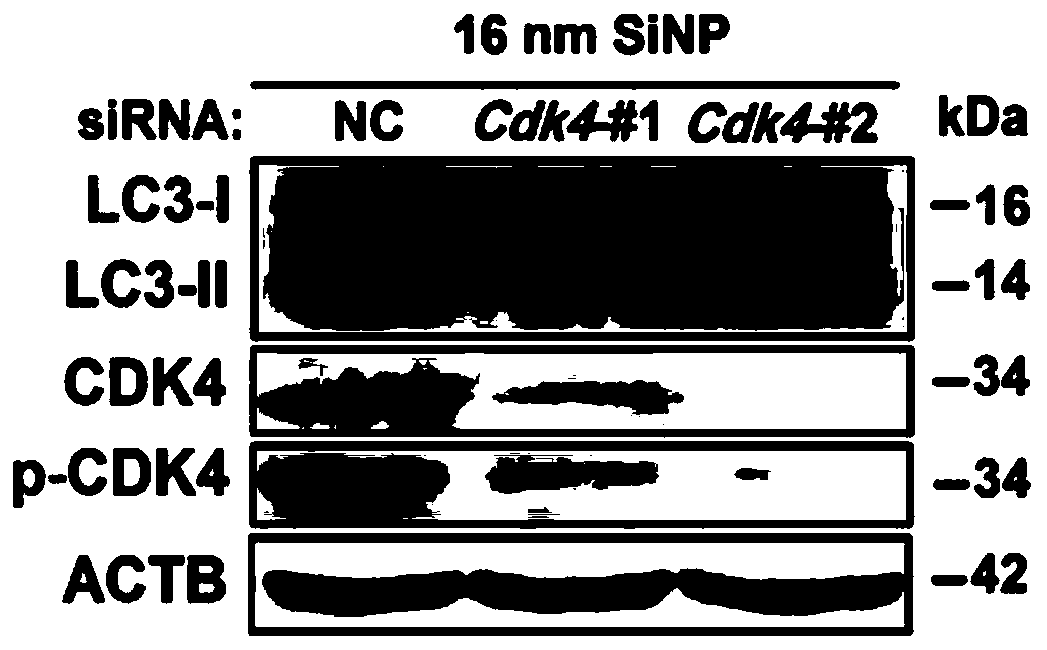
[0068] Subsequently, small molecule interfering RNA was designed for the above kinase genes, and the expression changes of the autophagy marker molecule LC3-II were detected by Western blotting to explore the effect of protein kinases on autophagy. At the same time, the GFP-LC3 cell line was used to detect the effect of protein kinase on the formation of autophagosomes. Such as image 3 , Figure 4 As shown, knocking down Cdk4 reduced the expression level of LC3-II protein induced by nanomaterials, and inhibited the accumulation of GFP-LC3 polysites induced by nanomaterials. Such as Figure 5 and Figure 6 As shown, knocking down Cdk7 reduces the expression of LC3-II protein induced by nanomaterials, and reduces the number of GFP-LC3 aggregation points induced by nanomaterials. It was confirmed that two protein kinases, Cdk4 and Cdk7, played important regulatory functions in nanomaterial-induced autophagy.
[0069] Based on the above results, it is proved that this method...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com