Method for predicting functional gene for regulating and controlling yeast autophagy
A technology of autophagy genes and gene regulation, applied in the field of bioinformatics, can solve the problems of long experiment cycle and high cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
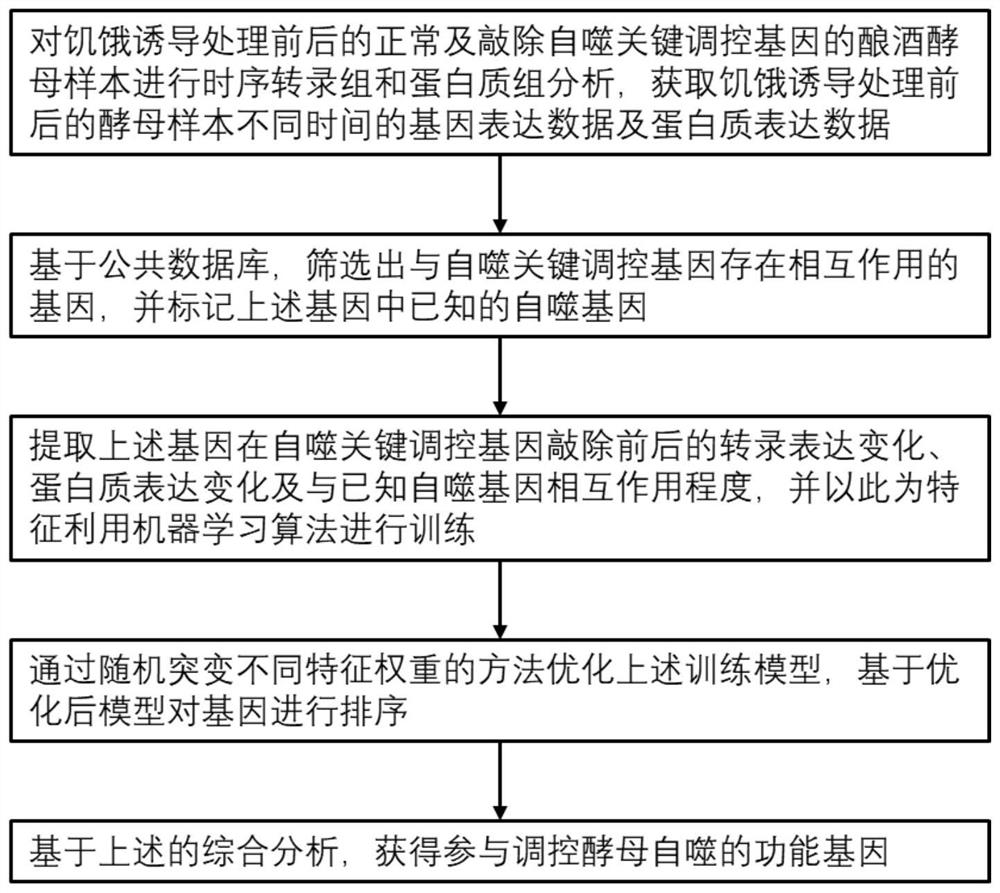
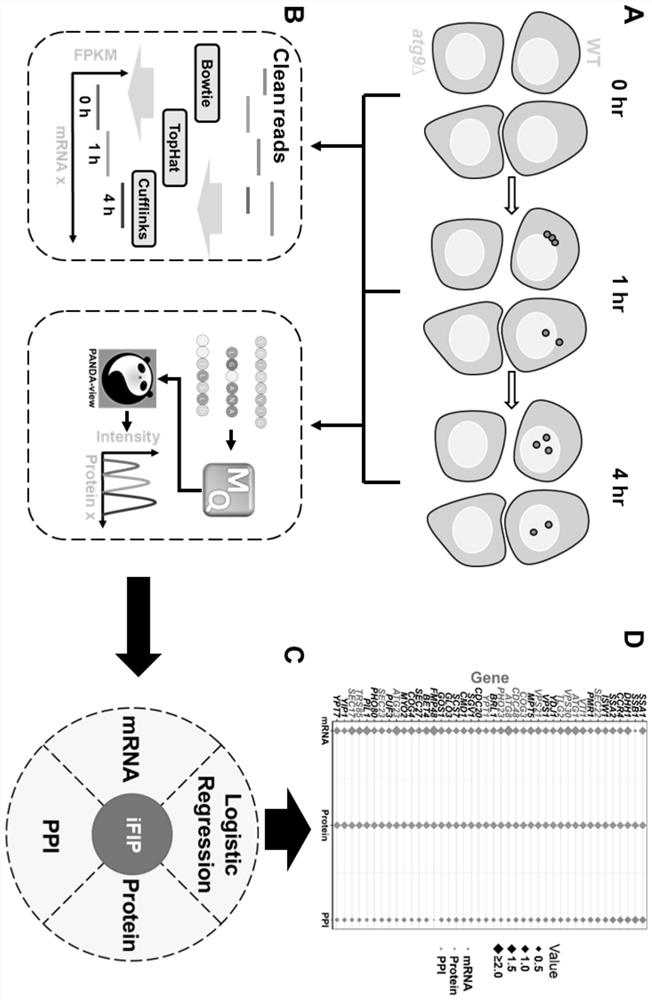
[0048] This example provides a method for predicting functional proteins involved in the regulation of autophagy based on multi-omics integration, such as figure 1 and figure 2 shown, including the following steps:
[0049] The wild-type (BY4741; MATa his3D leu2Dmet15D ura3D) and corresponding atg9 gene knockout yeast cells were treated with nitrogen source-depleted medium as an autophagy inducer for 0 hour, 1 hour, and 4 hours, respectively, and collected 3 hours after treatment Spot the yeast cell sample.
[0050] Using the second-generation gene sequencer HiSeq 4000system, RNA sequencing was performed on yeast cell samples before and after nitrogen-deficient medium treatment to obtain transcriptome data.
[0051] The Bowtie-Tophat-Cufflinks series software was used to analyze and process the transcriptome data to obtain quantitative information of genes.
[0052] Using liquid chromatography-mass spectrometry (LC-MS), the proteome analysis of yeast cell samples before an...
Embodiment 2
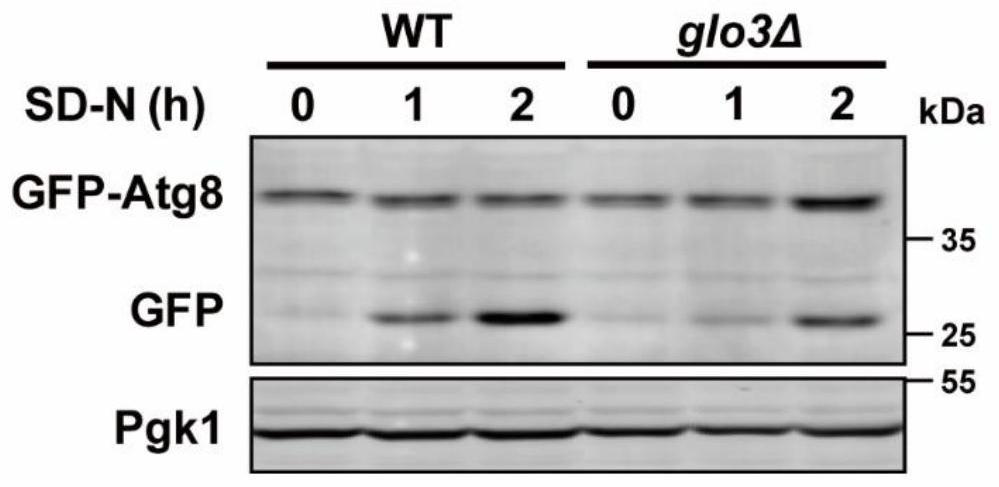
[0062] In order to verify whether the gene is involved in the regulation of autophagy, in this example, the potential genes involved in the regulation of autophagy, GLO3 and SCS7 obtained in the above-mentioned Example 1, were taken as examples, combined with the yeast gene knockout library, GFP was detected by Western blotting method -The splicing changes of Atg8 protein to explore the influence of predicted genes on autophagy in yeast cells. At the same time, according to the situation of the GFP-Atg8 protein entering the vacuole, the influence of the predicted gene on the autophagy activity of the cell was judged. like image 3 , Figure 4 As shown, knockdown of GLO3 can block the degree of cleavage of GFP-Atg8 induced by nitrogen source deprivation treatment and reduce the entry of GFP-Atg8 into vacuoles. like Figure 5 , Image 6 As shown, knockdown of SCS7 can reduce the cleavage of GFP-Atg8 induced by nitrogen source deprivation treatment and prevent GFP-Atg8 from e...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com