Polypeptide capable of targeting triple-negative breast cancer cells, and application thereof
A technology of recombining cells and amino acids, which is applied in general/multifunctional contrast agents, medical preparations with non-active ingredients, and medical preparations containing active ingredients, etc. Incomplete cells, etc., to achieve the effect of high specificity, affinity, and high specificity selection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
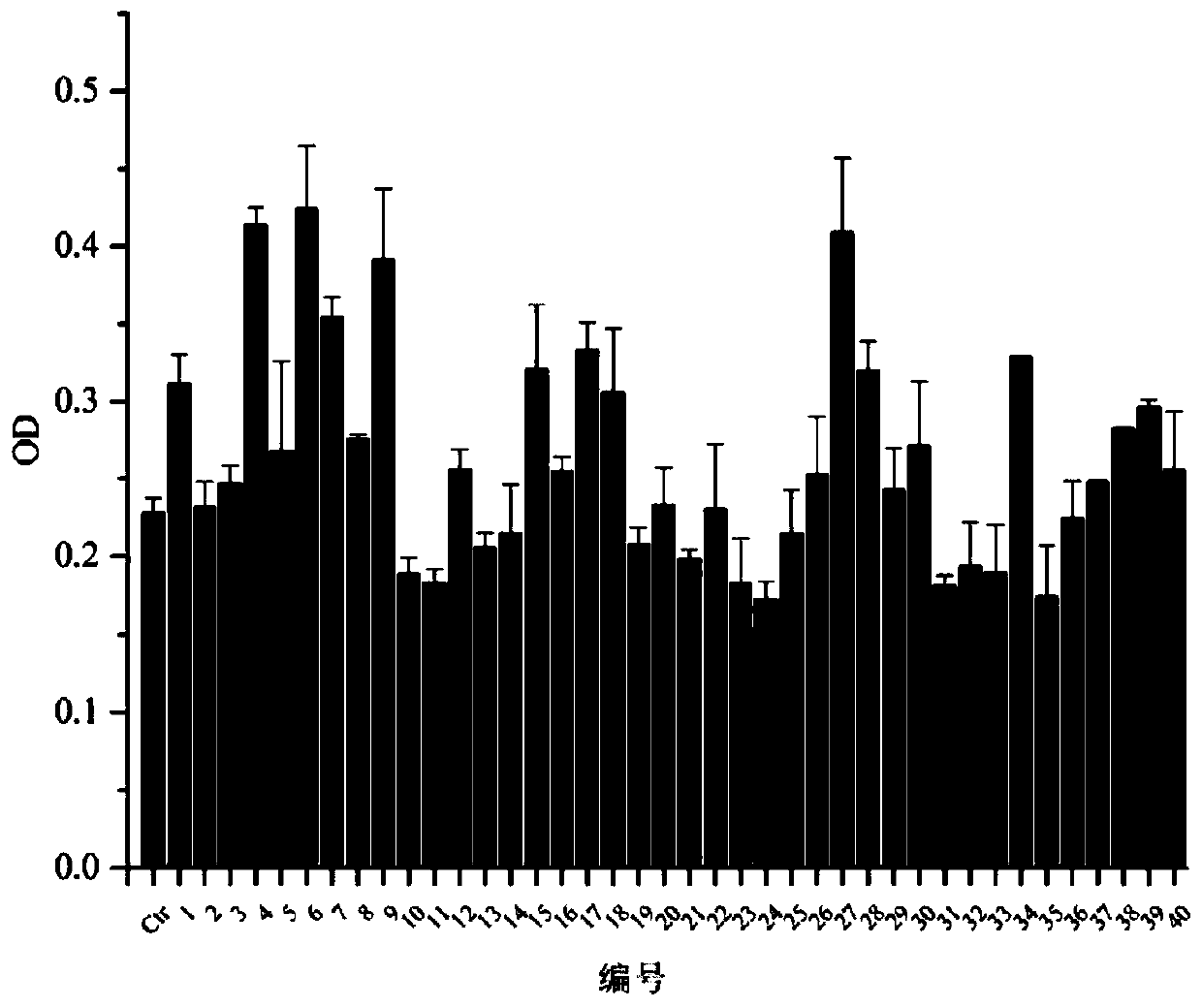
[0042] Screening Synthesis of Peptides
[0043] Cells were screened using the Negative- / Positive+ method using the phage peptide library. Among them, breast cancer cells of different types, including MDA-MB-453 and MCF-7, are used for negative screening cells to exclude polypeptides that can bind to these negative screening cells; triple negative screening cells are selected for positive screening. Breast cancer cell MDA-MB-231 is used to select polypeptides that can specifically bind to it. There were three rounds of screening. The screening steps are as follows:
[0044]1) MCF-7, MDA-MB-453, SK-BR3, BT-474, T47D were used as negative screening cells, triple-negative breast cancer cell MDA-MB-231 was used as positive screening cells and cultured in RPMI- 1640 medium.
[0045] 2) After negative screen cells were blocked with 0.5% BSA blocking solution for 1 h, the phage random dodecapeptide library (Ph.D.-12 phage display peptide library kit, New England Bio-LABS, USA) was...
Embodiment 2
[0065] Affinity and binding strength experiments
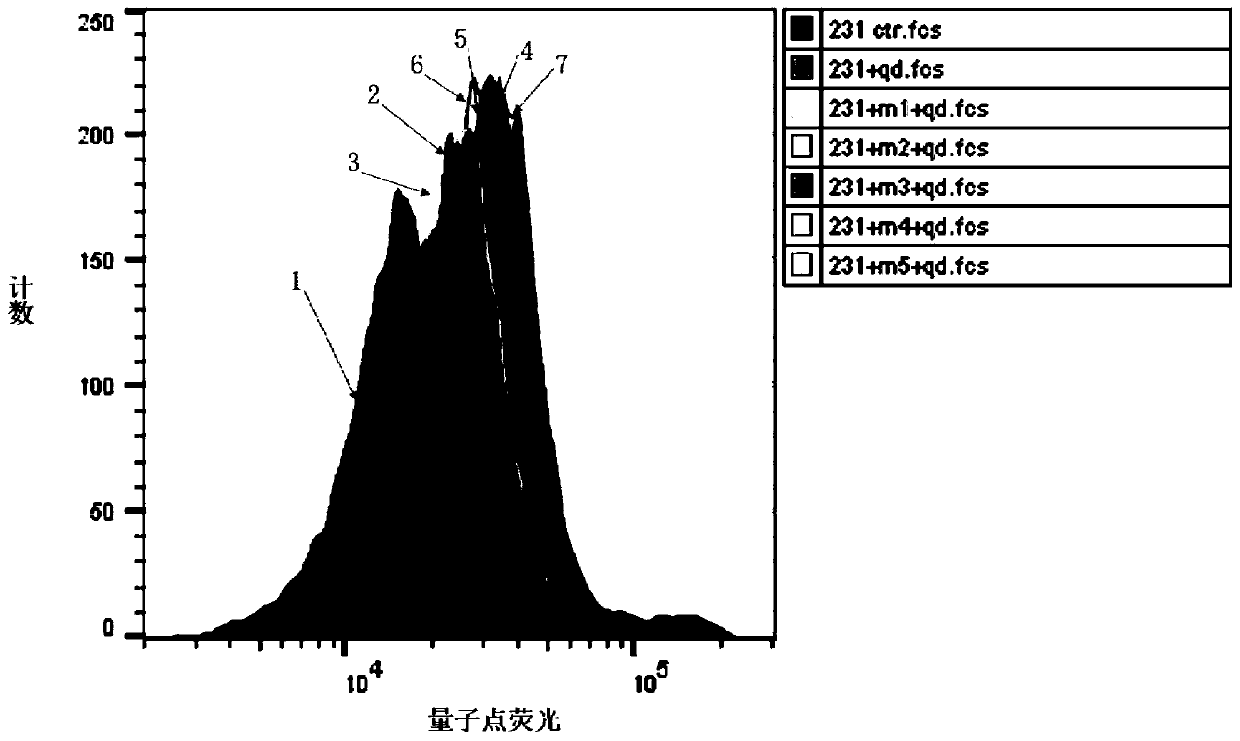
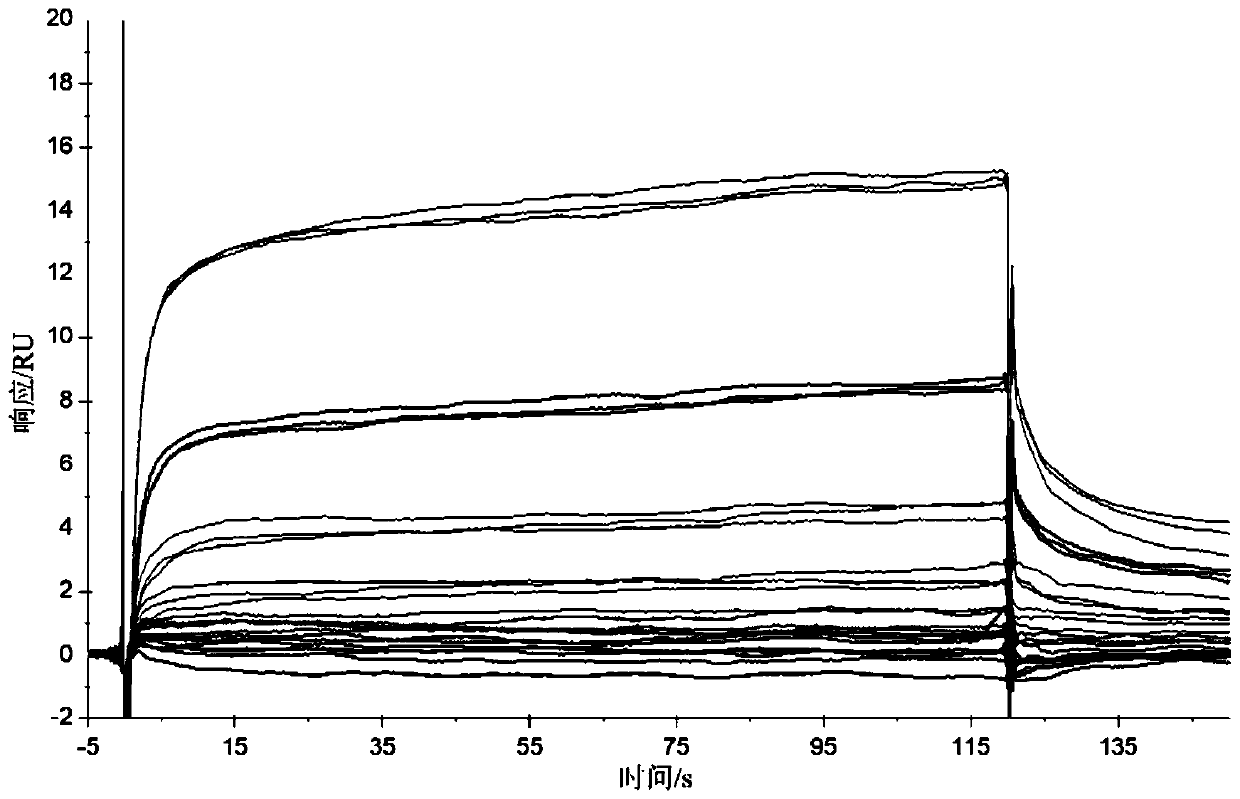
[0066] In this example, flow cytometry and surface plasmon resonance (SPR) were used to analyze the five polypeptides screened in Example 1 (the polypeptides of SEQ ID No.1 to SEQ ID No.5 are represented by polypeptides 1 to 5, respectively) Affinity and binding constant with triple-negative breast cancer cell MDA-MB-231, the specific steps are as follows:
[0067] a. After the five polypeptides were labeled with quantum dots, they were incubated with triple-negative breast cancer cells MDA-MB-231, and the fluorescence intensity was analyzed by flow cytometry to determine the affinity of the five polypeptides with triple-negative breast cancer cells.
[0068] b. Extract MDA-MB-231 membrane protein from triple-negative breast cancer cells.
[0069] c. The MDA-MB-231 membrane protein of triple-negative breast cancer cells was coupled to the SPR chip by using EDC / NHS.
[0070] d. The targeting polypeptides were formulated into ...
Embodiment 3
[0074] Fluorescent probe targeting triple-negative breast cancer cells
[0075] A fluorescent probe specifically targeting triple-negative breast cancer cells, comprising the polypeptide whose amino acid sequence is shown in SEQID No.5 in Example 1 and the fluorescent group FITC coupled to the peptide chain.
[0076] To detect the targeting specificity of the fluorescent probe, the specific steps are as follows:
[0077] 1) Synthesizing a polypeptide linked to a FITC fluorescent group;
[0078] 2) Inoculate triple-negative breast cancer cells MDA-MB-231 and SK-BR3 cells as a control group in a 24-well plate, and wait for the cells to grow to an appropriate density;
[0079] 3) Aspirate the medium, wash with PBS three times, each time for 3 minutes, add 500 μL of 4% paraformaldehyde solution to each well, fix at room temperature for 20 minutes, absorb the fixative, wash with PBS solution three times, each time for 3 minutes;
[0080] 4) Dilute the polypeptide at 1:500, add to...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com