Method for high-yield production of L-cysteine by metabolic engineering modification of corynebacterium glutamicum
A technology of Corynebacterium glutamicum and cysteine, which is applied in the fields of synthetic biology and metabolic engineering, and can solve the problems of difficulty in meeting industrial production needs and low yield.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
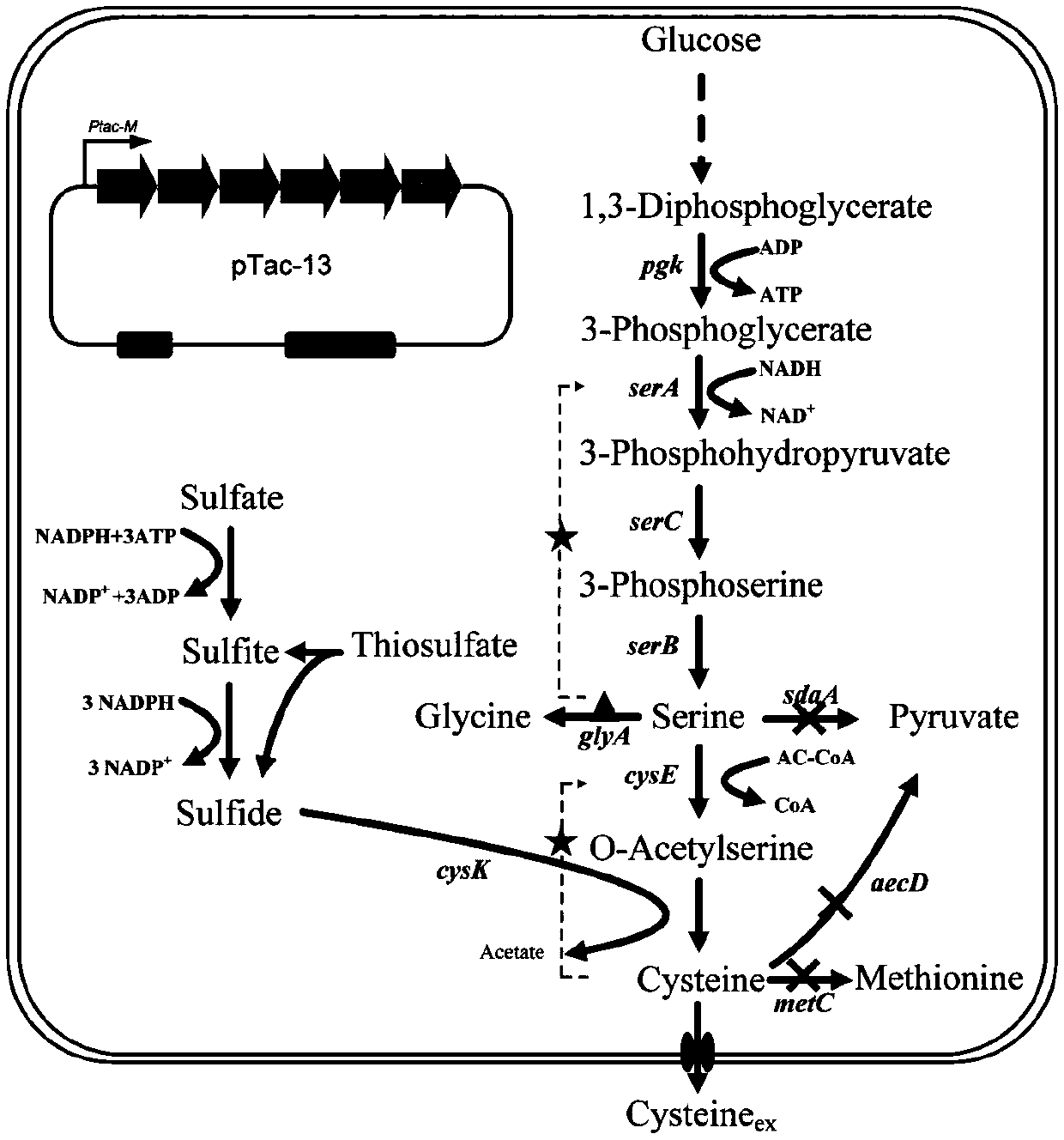
[0018] Example 1 Construction of Corynebacterium glutamicum L-cysteine desulfhydrylase gene aecD deletion strain
[0019] (1) Extraction of C. glutamicum ATCC13032 genome template
[0020] C. glutamicum ATCC13032 was cultured in LB medium (20-25g / L peptone, 10-20g / L yeast powder, 20-25g / L sodium chloride) at 30°C and 150-200rpm for 12-16h, then used genome extraction reagent Kit (Tiangen) to extract total DNA.
[0021] (2) Design primers aecD1-F / aecD1-R, aecD2-F / aecD2-R using the C. glutamicum ATCC13032 genome as a template, and their sequences are:
[0022] aecD 1-F: cgcggatcccgacagtcaatgcgatgcc;
[0023] aecD 1-R:acgaagaattttagaaggcctttccgcaacccacaaagg;
[0024] aecD 2-F: tgtgggttgcggaaaggccttctaaaattcttcgtgagg;
[0025] aecD 2-R: tgctctagaaacggccaacaccgtatcc.
[0026] Using the primers designed above, fragments aecD1 and aecD2 were obtained by PCR. Then obtain the PCR products aecD1 and aecD2 as templates, use aecD1-F / aecD2-R as primers, obtain the fusion fragment a...
Embodiment 2
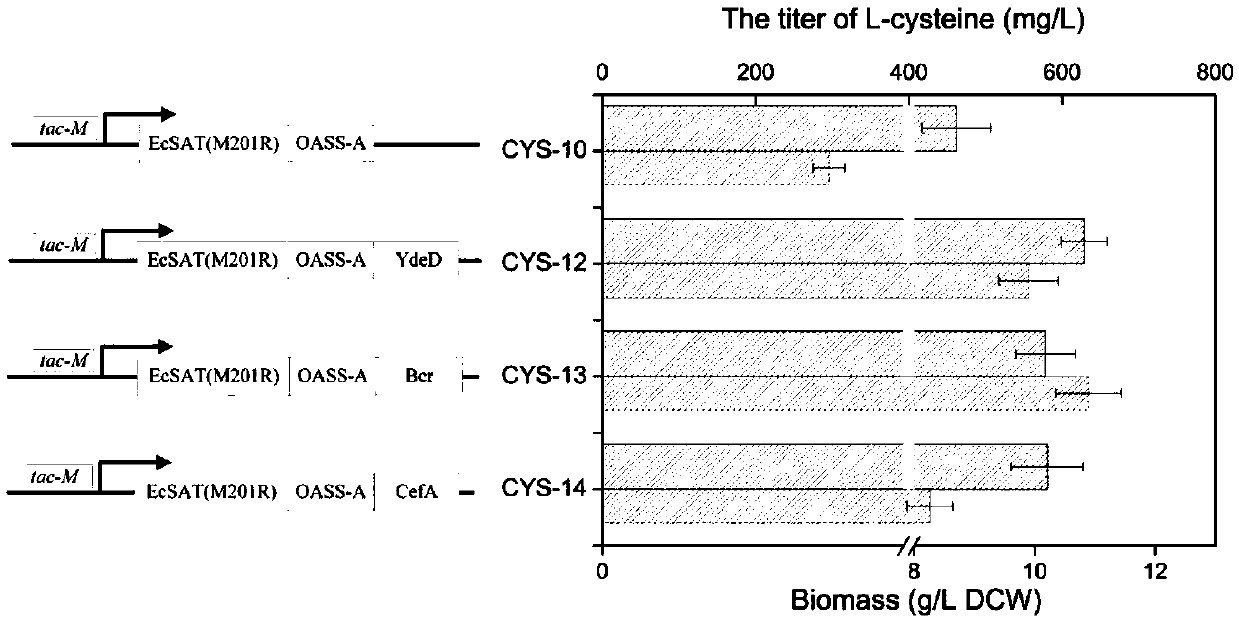
[0036] Example 2 Construction of overexpressed self-serine transacetylase gene cysE strain
[0037] (1) Design primers CgcysE-F / CgcysE-R using the C. glutamicum ATCC13032 genome as a template, and its sequence is:
[0038] CgcysE-F: tccgagctcaaaggaggacaaccatgatccgtgaagatctcgca;
[0039] Cgcys E-R: cggggtaccatagggcgctaactgttcttaaatg.
[0040] Using the above primers, the fragment CgcysE was obtained by PCR. Then, after double digestion with SacI and KpnI, it was connected to the vector pTrcmob to obtain the plasmid pTrc-1.
[0041] PCR system: ddH 2 O 30 μL, template 1 μL, Fastpfu buffer 10 μL, Fastpfu 1 μL, primer 1 1.5 μL, primer 21.5 μL, dNTP 5 μL.
[0042] PCR conditions: Step 1 at 94°C for 4min, Step 2 at 94°C for 30s, Step 3 at 60°C for 30s, Step 4 at 72°C for 1min, Step 5 at 72°C for 10min, and Step 6 at 4°C.
[0043] Double digestion system: gene aecD or plasmid pCRD206 20 μL, BamHI 1 μL, XbaI 1 μL, 10×buffer 5 μL, ddH 2 O 23μL, react at 37℃ for 1h.
[0044] Liga...
Embodiment 3
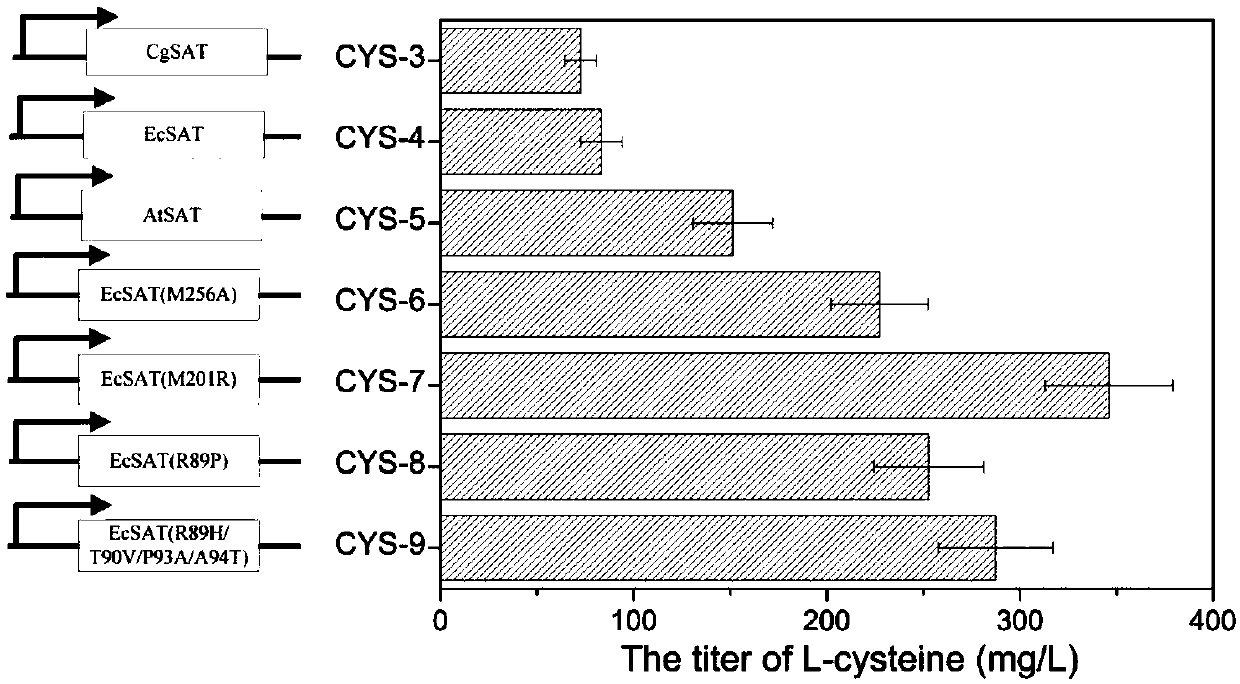
[0047] Example 3 Exploring the influence of serine transacetylase gene cysE from different sources on L-cysteine
[0048] (1) Extract the E.coli MG1655 genome template
[0049] E.coli MG1655 was cultured in LB medium (20-25g / L peptone, 10-20g / L yeast powder, 20-25g / L sodium chloride) at 37°C and 150-200rpm for 12-16h, then the genome extraction reagent was used Kit (Tiangen) to extract total DNA.
[0050] (2) Design primers EccysE-F / EccysE-R using the E.coli MG1655 genome as a template, the sequence of which is:
[0051] EccysE-F: tccgagctcaaaggaggacaacccaatgtcgtgtgaagaactgga;
[0052] Eccys E-R: cggggtaccatgattacatcgcatccgg.
[0053] Using the primers designed above, the fragment CgcysE was obtained by PCR. Then, after double digestion with SacI and KpnI, it was connected to the vector pTrcmob to obtain the plasmid pTrc-2.
[0054] (3) Based on the codon usage preference of Corynebacterium glutamicum, the codon-optimized Arabidopsis cysE gene, whose sequence is shown in ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com