Prokaryotic acetylation site prediction method based on information fusion and deep learning
A prokaryotic and deep learning technology, applied in the field of bioinformatics, can solve the problems of underestimating the importance of multi-information fusion, limited prediction accuracy, and failure to integrate multiple feature information
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0054] In order to make the object, technical solution and advantages of the present invention clearer, the invention will be further described in detail below in conjunction with the accompanying drawings and embodiments. It should be understood that the specific embodiments described here are only used to explain the present invention, not to limit the present invention.
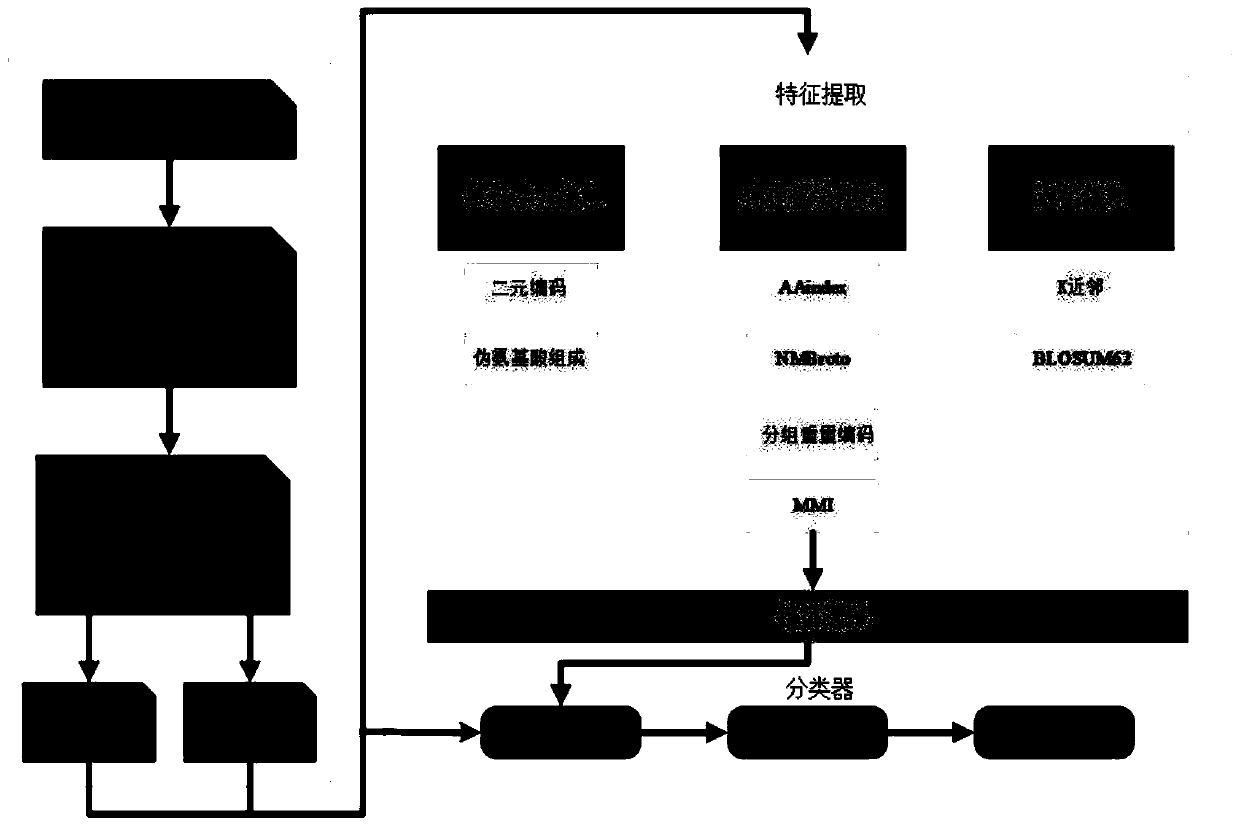
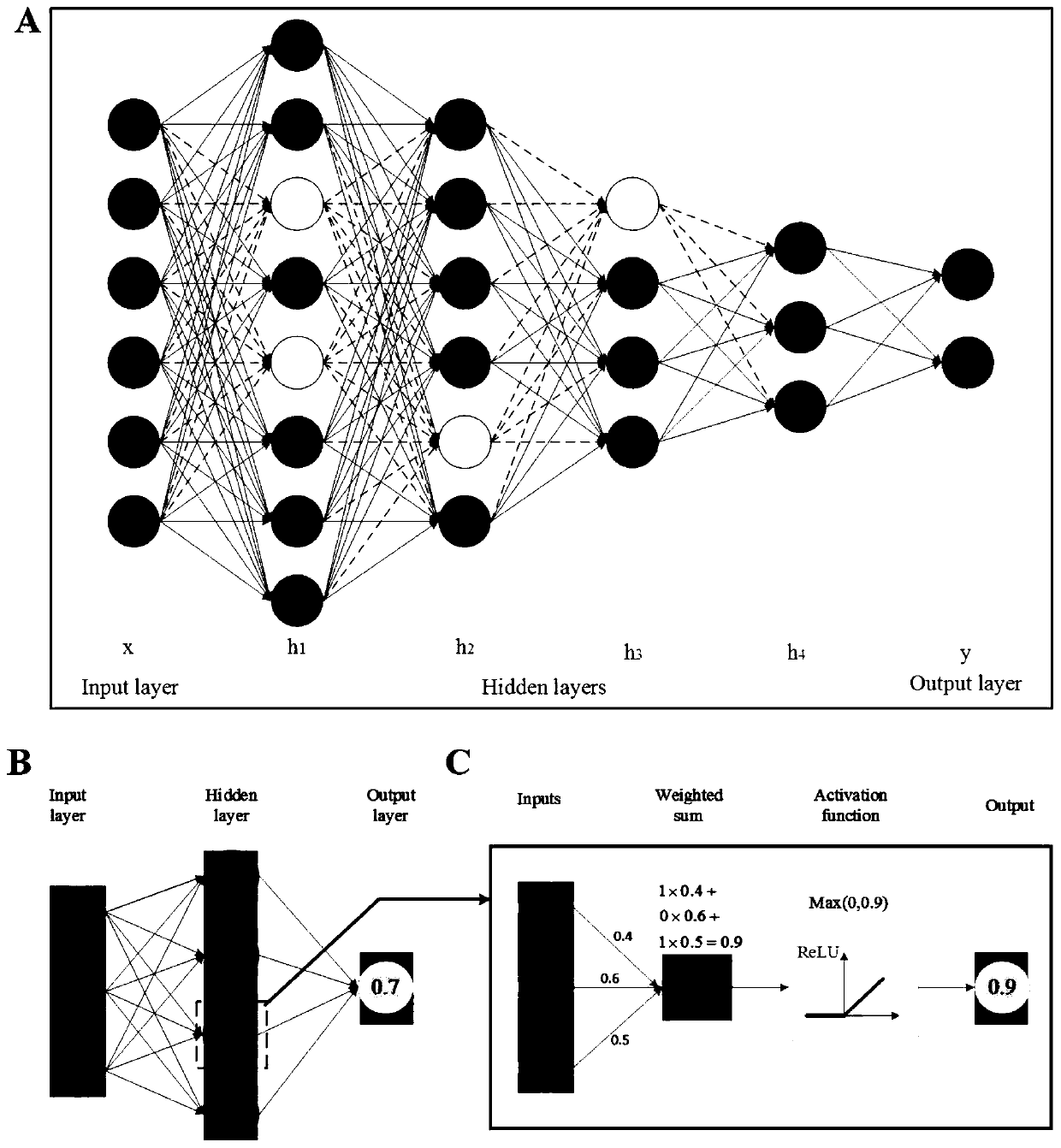
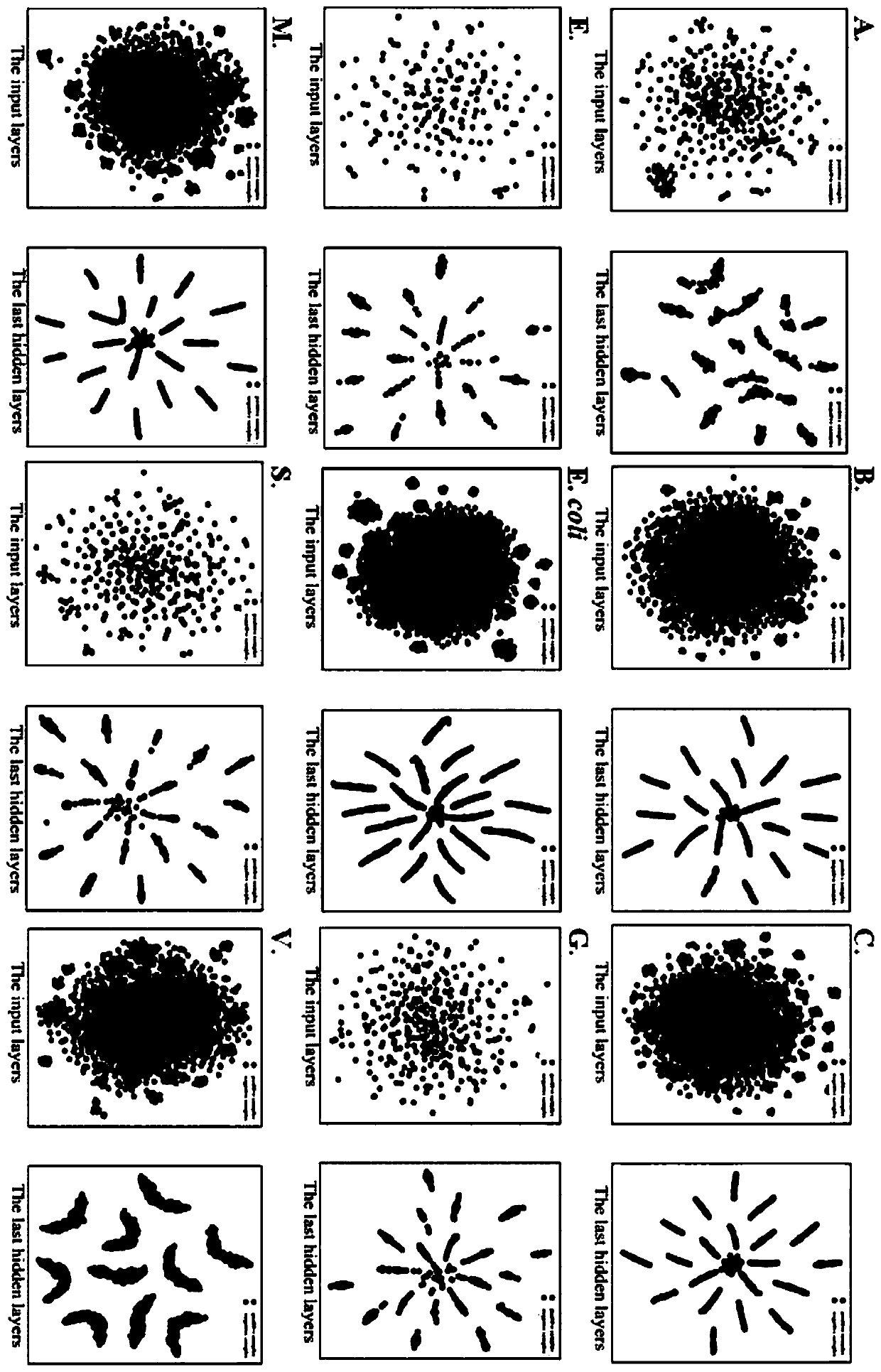
[0055] A prokaryotic acetylation site prediction method based on information fusion and deep learning, such as figure 1 shown, including the following steps:
[0056] 1) Collect acetylation modification site information: Obtain 9 prokaryotic acetylation site data sets from relevant literature, and generate category labels corresponding to positive and negative samples.
[0057] 1-1) The data sets of lysine acetylation sites of 9 categories of prokaryotes were constructed, namely E.coli, S.typhimurium, Bacillus subtilis (B.subtilis), Vibrio parahemolvticus (V.parahemolvticus), Mycobacterium tuberculosis (...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com