Esterases and coding gene and application thereof
A technology of esterase and gene, which is applied in the field of esterase and its coding gene and application, can solve the problems of not being environmentally friendly, relying on toxic substances, special reaction conditions, etc., and achieve high enzyme activity, high tolerance, and huge application potential Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0063] Example 1: Acquisition and sequence analysis of the gene sequence encoding esterase E31-4922 derived from Antarctic soil
[0064] The source of the strain: Escherichia coli EPI300 clone E31-4922 in the metagenomic library of soil samples at site E2 near the Great Wall Station in Antarctica.
[0065] Specific steps are as follows:
[0066] 1.1 Construction of subcloning library
[0067] Use the BAC / PAC DNA extraction kit from OMEGA Company to extract the large fragment plasmid fosmid in E. coli EPI300 clone E31-4922 according to its instructions. Then the fosmid extracted was partially digested with restriction endonuclease Sau3AI (purchased from Fermentas Company) to obtain a DNA fragment of 2,000-5,000bp, which was connected to the pUC19 plasmid (purchased from BamHI) digested and dephosphorylated by BamHI. from NEB Corporation). Electroporate E.coli Top10 competent cells with the ligation reaction solution, coat LB solid plate containing 100 μg / ml ampicillin and 1%...
Embodiment 2
[0072] Example 2: Cloning, heterologous expression and separation and purification of esterase
[0073] 2.1 Using PCR to amplify the gene sequence
[0074] (1) Design two specific primers according to the gene E31-4922 sequence:
[0075] 4922F: aagaaggaga tatacatatg agctacccgg ctatcggtta ctg (SEQ ID NO. 3);
[0076] 4922R: tcgagtgcgg ccgcaagctt caggtgagaa cggctttccg agc (SEQ ID NO. 4);
[0077] Primers were synthesized by Jinan Boshang Biotechnology Co., Ltd.
[0078] (2) Using 4922F and 4922R as primers, using the fosmid where the gene E31-4922 is located as a template, amplify the target gene fragment with FastPfuDNA polymerase (purchased from Transgen);
[0079] The PCR reaction conditions were: pre-denaturation at 95°C for 2 min; then denaturation at 95°C for 20 sec, annealing at 50°C for 20 sec, extension at 72°C for 1 min, and after 30 cycles; extension at 72°C for 10 min.
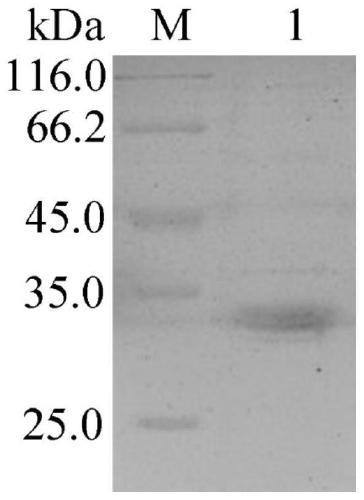
[0080] (3) 1 wt% agarose gel electrophoresis was performed on the PCR amplification product, ...
Embodiment 3
[0103] Embodiment 3: the property determination of Antarctic esterase E31-4922
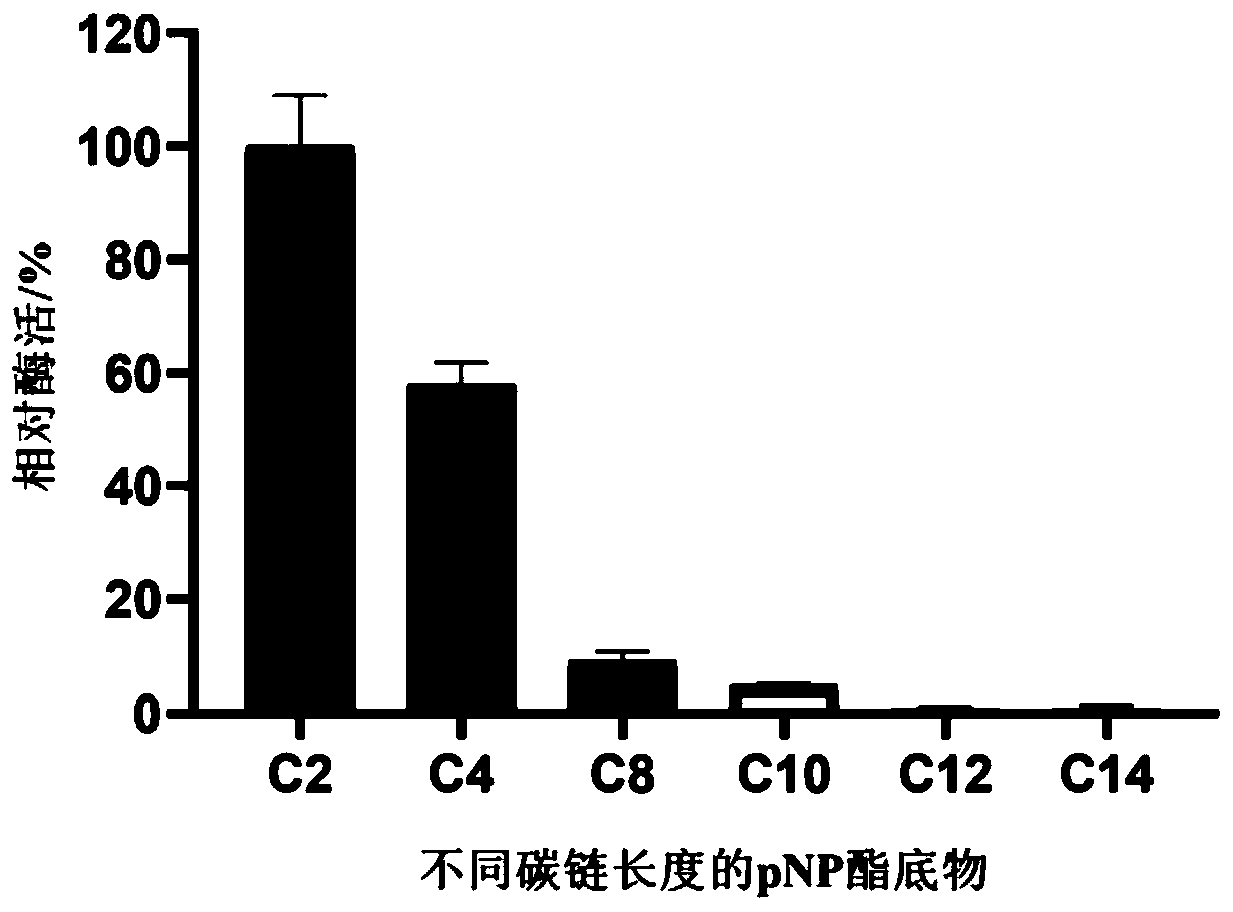
[0104] 3.1 Substrate specificity analysis
[0105] pNP ester substrates C2, C4, C8, C10, C12, and C14 (purchased from Sigma) with different carbon chain lengths were prepared with isopropanol.
[0106] The standard response is:
[0107] 20μl of 10mM pNPC4 substrate and 960μl of 50mM Tris-HCl (pH 8.0) mixture was preheated at 40°C for 3 minutes, then 20μl of the diluted enzyme solution was added and reacted at 40°C for 10min, and 100μl of 20wt% SDS (dodecyl Sodium sulfate) to stop the reaction, measure the OD405 value. The reaction without enzyme solution was used as blank control. The standard curve was drawn with different concentrations of pNP (purchased from Sigma).
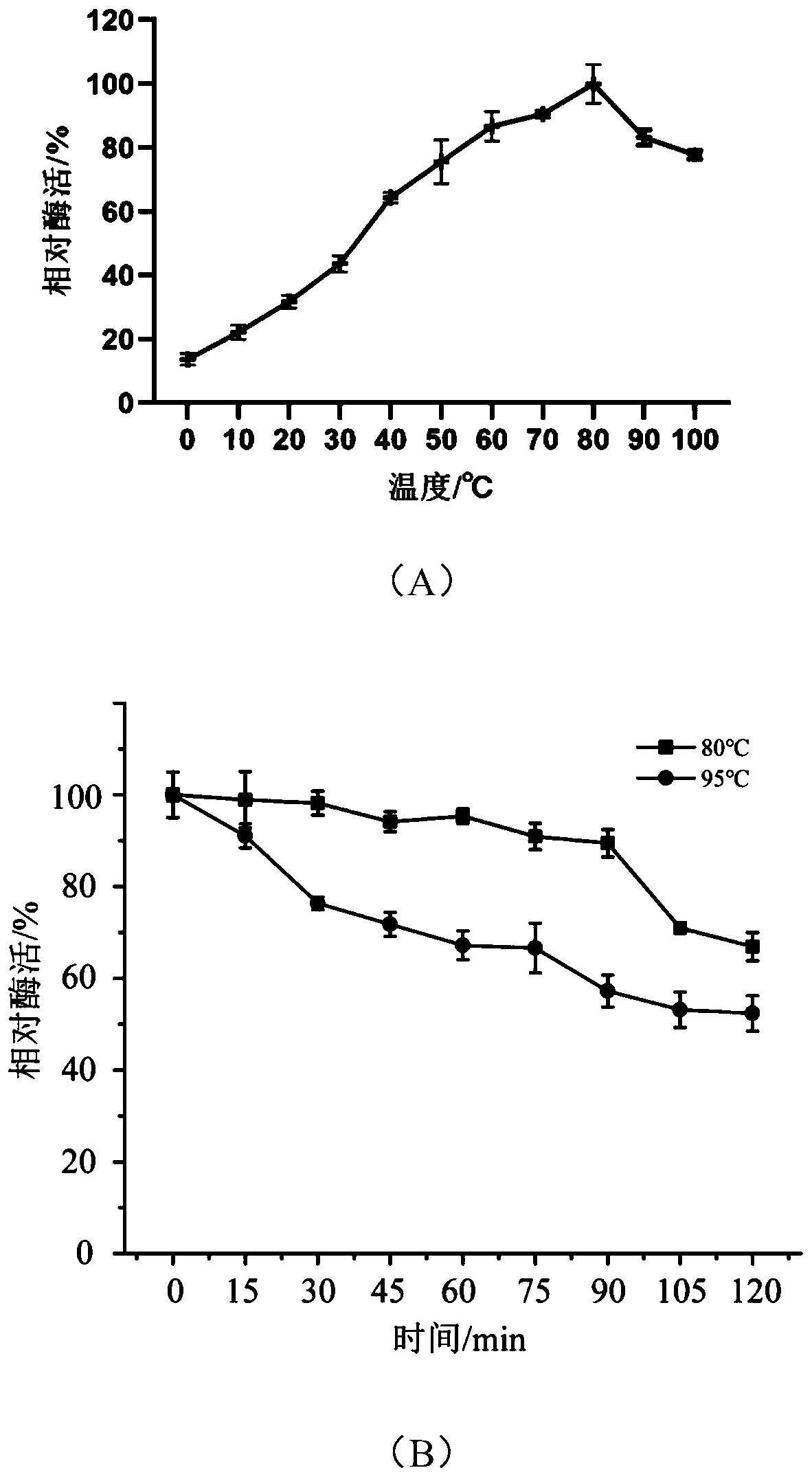
[0108] Enzyme activity is defined as the amount of enzyme required to catalyze the hydrolysis of pNP ester substrate to produce 1 μM pNP per minute at a certain temperature, which is one enzyme activity unit (U). The results ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com