Primer, probe and kit for detecting novel coronavirus
A technology of coronaviruses and kits, applied in biochemical equipment and methods, measurement/inspection of microorganisms, resistance to vector-borne diseases, etc., to achieve the effects of less templates, overcoming false negatives, and improving accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0059] The design of embodiment 1, specific primer and probe
[0060] In the design of primer probes, it is necessary to select regions that are conserved within the group and specific among groups for design, so as to avoid false negatives caused by missed detection, or false positives caused by poor specificity.
[0061] Through sequence comparison and literature search, the present invention finds that the structural genes S, M, ORF1ab, E and N of 2019-nCoV are highly conserved and can be used as target regions for the identification and detection of new coronaviruses.
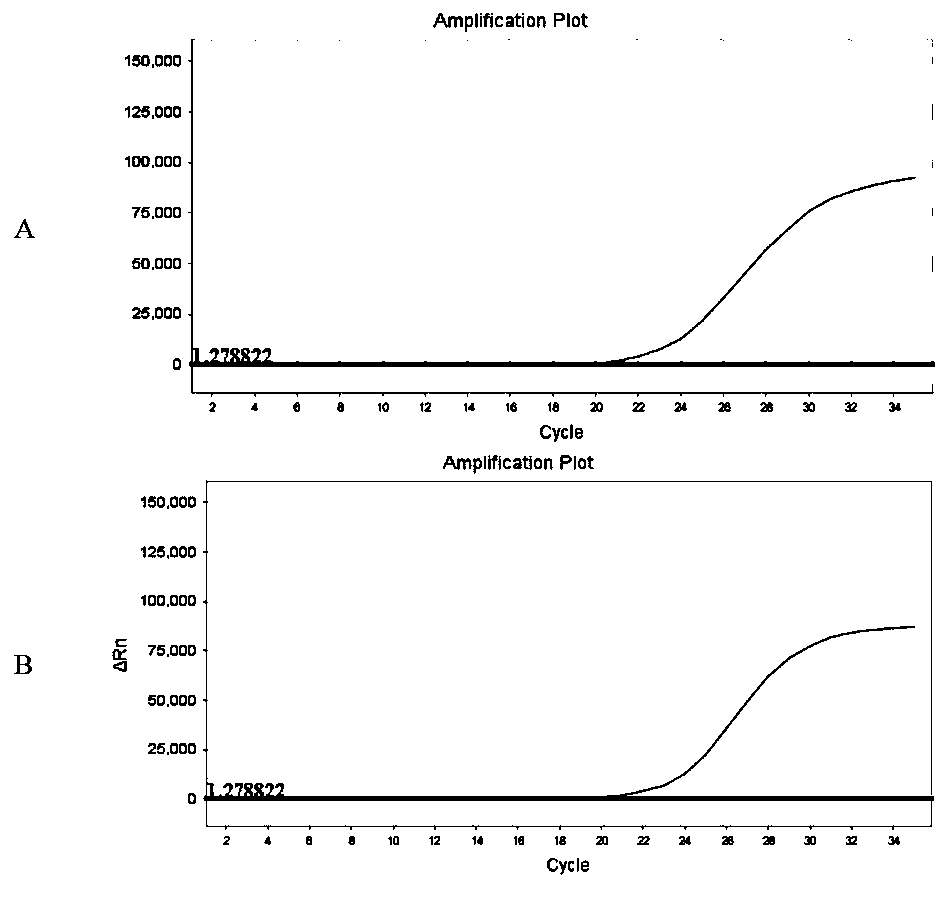
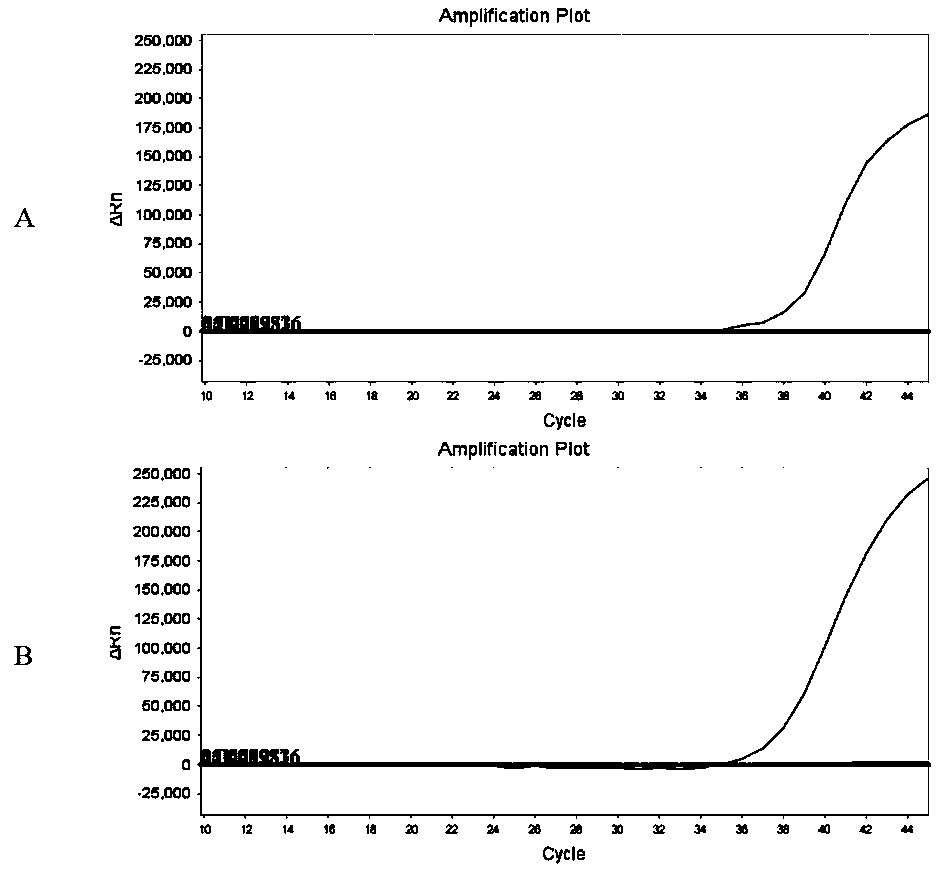
[0062] In this example, the sequences of endemic human coronavirus (HKU1, OC43, 229E and NL63), SARS coronavirus, MERS coronavirus, bat coronavirus (MG772933_Bat_bat_SL_CoVZC45), SARS-like virus (MG772934_Bat_SARS_like_bat_SL_CoVZXC21) were selected for comparison, and the results are as follows figure 1 shown. Open reading frame 1ab (ORF1ab), nucleocapsid protein (N), envelope protein (E), surface glycopr...
Embodiment 2
[0113] Embodiment 2, comparative analysis of specific primers and probes of the present invention and CDC recommended sequences
[0114] In order to evaluate the preferred primers and probes of Example 1, the sequence recommended by CDC (Center for Disease Control and Prevention) was used as a comparison sequence for detection evaluation.
[0115] China CDC does not recommend primer and probe sequences for E gene, S gene and M gene, but only recommends primer and probe sequences for ORF1ab and N genes. The recommended primer and probe sequences for ORF1ab and N regions are as follows:
[0116] ORF1ab-F (SEQ ID NO: 16): 5'-CCCTGTGGGTTTTACACTTAA-3';
[0117] ORF1ab-R (SEQ ID NO: 17): 5'-ACGATTGTGCATCAGCTGA-3';
[0118] ORF1ab-P (SEQ ID NO: 18): 5'-CCGTCTGCGGTATGTGGAAAGGTTATGG-3';
[0119] N-F (SEQ ID NO:31): 5'-GGGGAACTTCTCCTGCTAGAAT-3';
[0120] N-R (SEQ ID NO: 32): 5'-CAGACATTTTGCTCTCAAGCTG-3';
[0121] N-P (SEQ ID NO: 33): 5'-TTGCTGCTGCTTGACAGATT-3'.
[0122] The US CDC ...
Embodiment 3
[0129] The optimization experiment of embodiment 3, RT-PCR method
[0130] There are two methods for detection of viral RNA based on fluorescent RT-PCR, that is, the two-step method of performing reverse transcription into cDNA first, and then performing PCR, and the one-step method system of simultaneously performing RT-PCR in the same system. Both have pros and cons. Using the two-step system, the cDNA obtained by reverse transcription is easy to preserve; while the one-step method is fast, simple, and reduces the chance of contamination. For multiplex PCR, interference between primers and probes needs to be considered.
[0131] The invention compares the detection effects of the two reaction systems, first evaluates the detection effect of a single pair of primers and probes, and then conducts a combination test of the synthesized sequences according to the reaction tubes.
[0132] In this example, Takara RNA PCR Kit (RR019A) and Takara Ex TAQ HS (RR390L) were used in the...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Copy number | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com