Device and method for extracting total DNA of matrix-attached microorganisms
A microorganism and matrix technology, applied in the field of molecular biology, can solve the problems of tedious post-processing steps, loss of low-abundance microorganisms, easy to contaminate samples, etc., to improve typicality and representativeness, the total band is bright and clear, and overcome sampling difficult effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
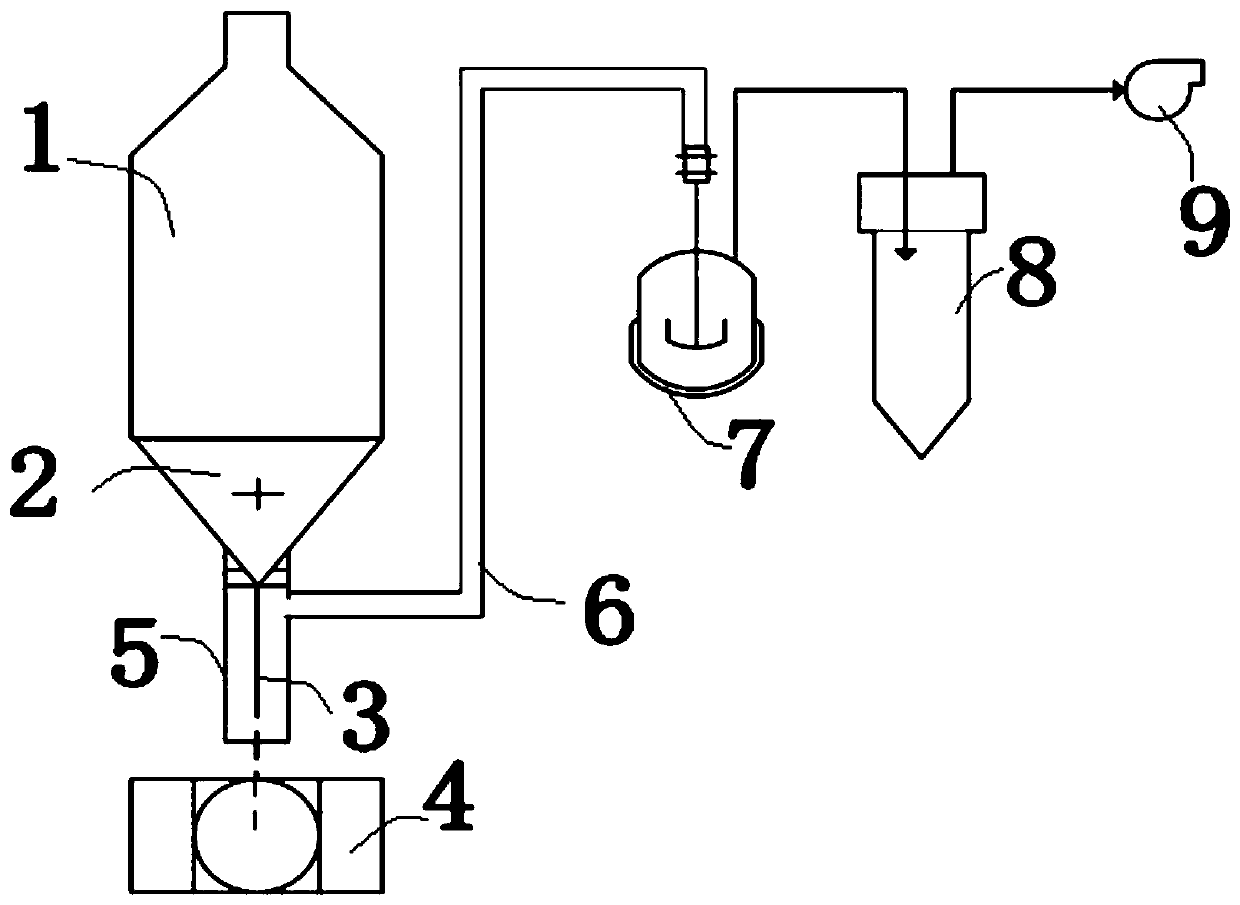
[0054] The device of the present invention for extracting substrate-attached total DNA of microorganisms, such as figure 1As shown, it includes a pressure-controlled flushing device 2, a negative pressure device and a collection device 8. The outlet end of the pressure-controlled flushing device 2 is provided with a flushing pipeline 3, and the pressure-controlled flushing device 2 is used to control the water outlet of the flushing pipeline 3. pressure, the negative pressure device includes a negative pressure pipeline and a negative pressure pump 9, the negative pressure pipeline includes a first negative pressure pipeline 5 and a second negative pressure pipeline 6, and the first negative pressure pipeline One end of 5 is sealed and connected with the pressure-controlled flushing device 2, and the other end is set as an open end; the flushing pipeline 3 is nested inside the first negative pressure pipeline 5, and the water outlet end of the flushing pipeline 3 is located at ...
Embodiment 2
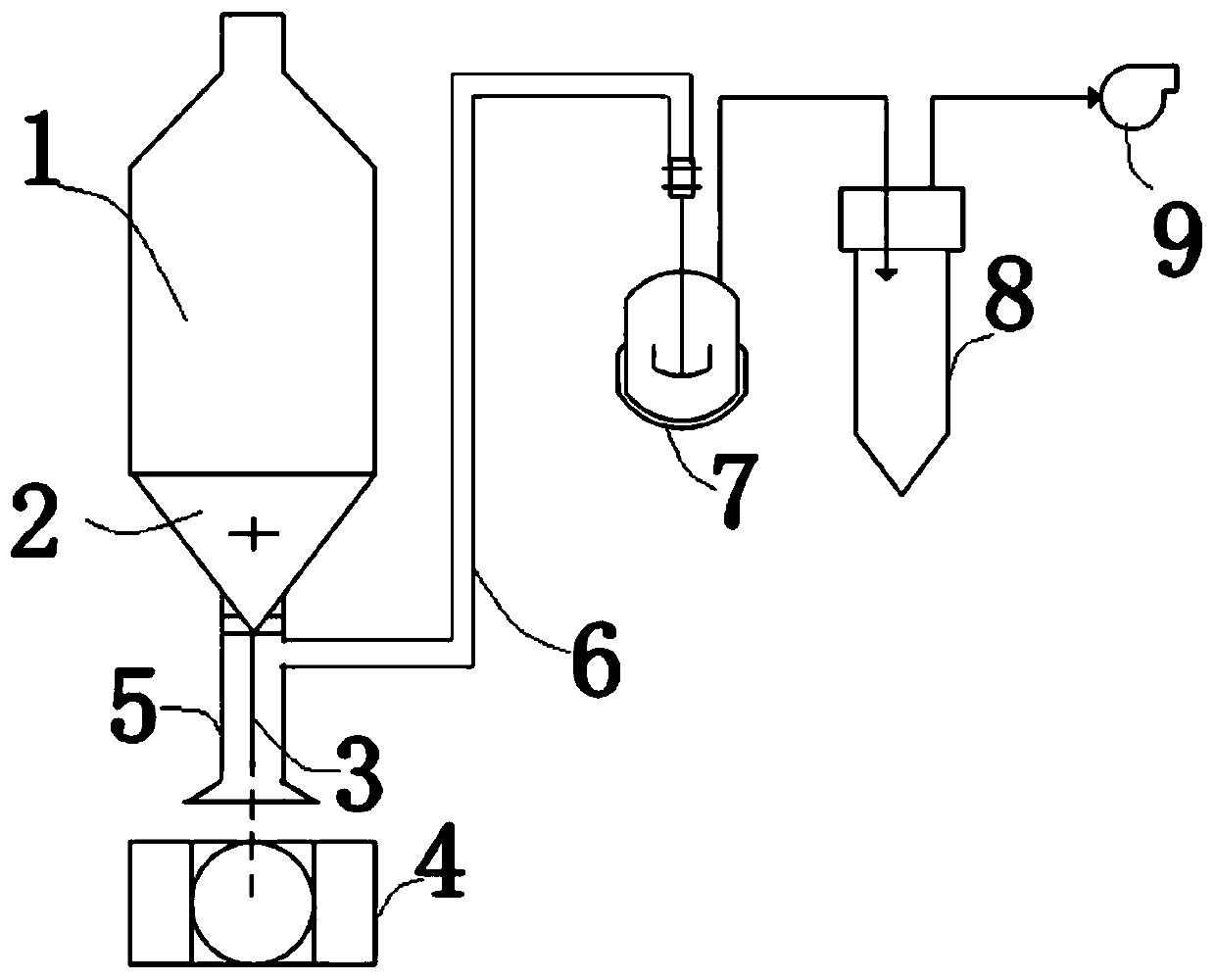
[0077] In order to meet the needs of more accurate sampling, the open end of the first negative pressure pipeline 5 in the device of the present invention can be connected to ports of different shapes to adapt to different actual situations, such as figure 2 As shown, the open end can be connected to a tapered port to ensure that the suspension eluate sputtered out after washing under different conditions can be collected more completely.
[0078] This embodiment also provides a DNA extraction method using the device of this embodiment. The method is aimed at the super acidic circulating fluid conditions of a flue gas desulfurization and denitrification biofilm packed tower described in Chinese Patent Publication No. CN109569270B The extraction and determination of bacterial total DNA and 16S rDNA attached to the filler, and the extraction and determination of total fungal DNA and iTS at the same time, the steps are as follows:
[0079] Step 1, collection of functional flora ...
Embodiment 3
[0083] In this example, the device of Example 1 is used to extract the total DNA of fungi attached to the surface of the filler in a denitrification biofilm filler reactor, and the steps are as follows:
[0084] Step 1. Collection of functional flora samples: From the denitrification biofilm filter filled with ceramsite fillers, take about 0.5L of ceramsite fillers with good microbial growth status and place them in a 1L beaker. In this example, ceramsite The diameter of the filler is 10-30mm, and the bulk density is 300kg / m 3 , specific surface area 230~300m 2 / m 3 , Porosity 53%. Put 100mL of the prepared phosphate buffer solution of pH=7 into a water pressure container, and use a pressure of 60MPa to flush the packing to wash down the bacterial cells attached to the packing, and then enter the centrifuge tube. Remove two 50mL centrifuge tubes containing the microbial suspension from the device one after another, and centrifuge at a speed of 5,000×g for 2 minutes until th...
PUM
| Property | Measurement | Unit |
|---|---|---|
| The inside diameter of | aaaaa | aaaaa |
| Diameter | aaaaa | aaaaa |
| Bulk density | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com