Primer for detecting sulfate reducing bacteria in jet fuel, application and detection method
A detection method and jet fuel technology, which are applied in biochemical equipment and methods, microbial determination/inspection, DNA/RNA fragments, etc. very good effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0042] Example 1: Primer Design and Synthesis
[0043] According to the dsrB gene sequence of sulfate-reducing bacteria, a set of specific primers were designed, including outer primers (F3, B3), inner primers (FIP, BIP), and a loop primer (LB, LF). The 5' end of LF was labeled with fluorescein isothiocyanate (FITC) as a probe. The primer sequences are shown in Table 1,
[0044] Table 1 Primers for sulfate-reducing bacteria
[0045]
[0046] According to the primer sequences in Table 1, LAMP primers were synthesized in Dalian Bao Biology Co., Ltd.
example 2
[0047] Example 2: Optimization of reaction conditions for the amplification of sulfate-reducing bacteria
[0048] The amplification conditions were optimized using the genomic DNA of the sulfate-reducing bacteria Desulfotomaculum ruminis as a template. The concentrations of primers prepared during LAMP reaction were: 0.2 μmol / L for F3 and B3, 1.6 μmol / L for FIP and BIP, and 0.8 μmol / L for LF and LB. LAMP premix is 20mmol / L Tris-HCl (pH 8.8), 10mmol / L KCl, 8mmol / L MgSO 4 , 10mmol / L (NH4) 2 SO 4 , 0.1% Tween 20, 0.8mol / L Betaine, 1.4mmol / L dNTPs, 8U / μL Bst DNA polymerase. The LAMP reaction system of sulfate-reducing bacteria is shown in Table 2.
[0049] Table 2 LAMP reaction system of sulfate-reducing bacteria
[0050]
[0051] In order to obtain the best LAMP amplification efficiency and comprehensively consider the applicable conditions of LFD, the primers provided by the present invention and the LAMP reaction system were used to perform LAMP amplification reaction...
example 3
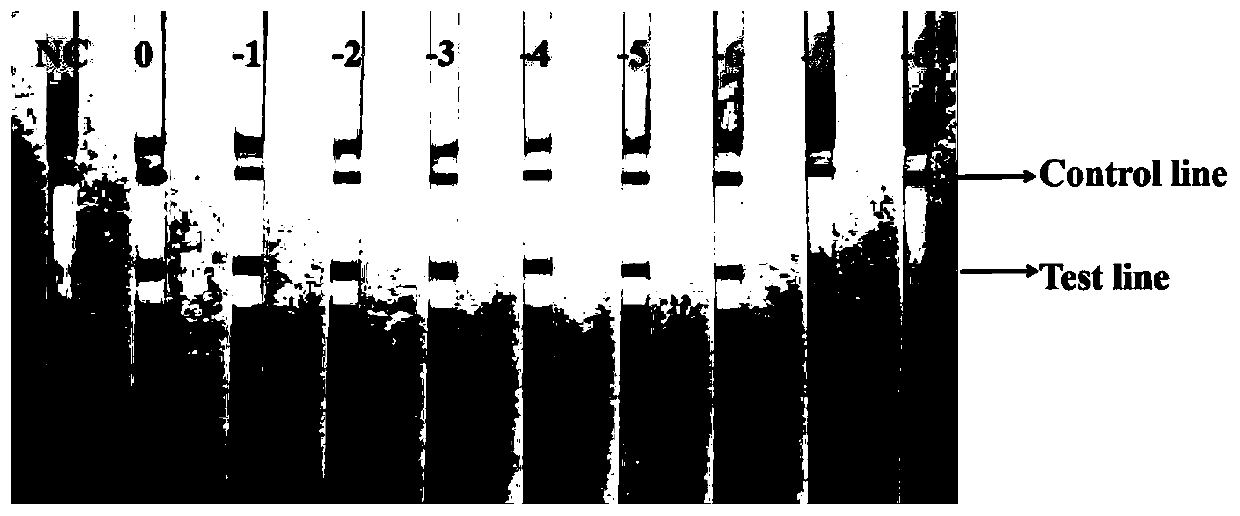
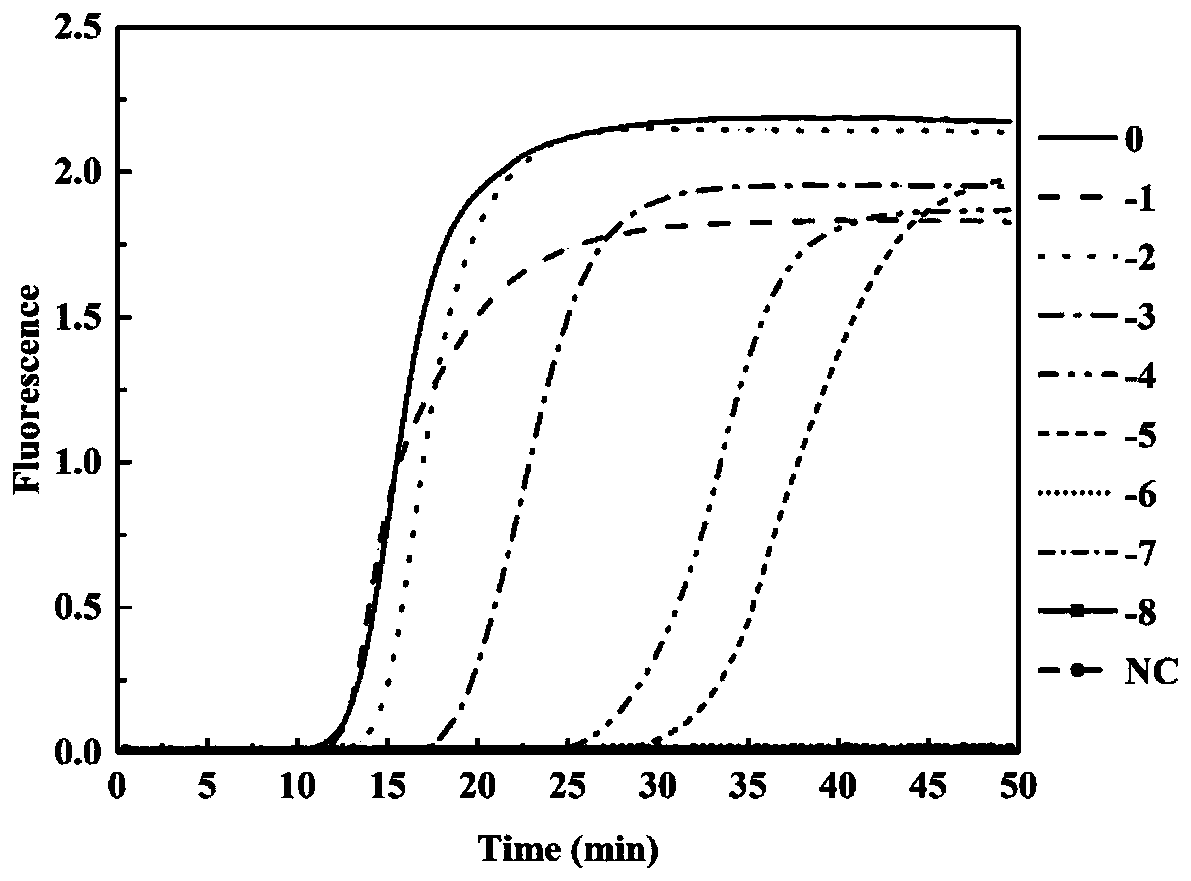
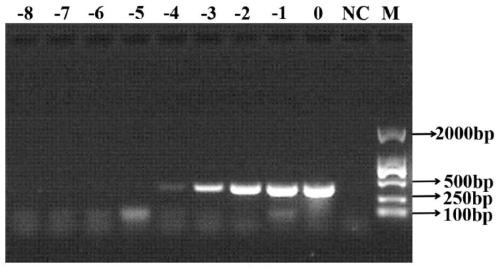
[0053] Example 3 uses the LAMP-LFD technique of the present invention to analyze the detection sensitivity of sulfate-reducing bacteria.
[0054] 1. Using the extracted Desulfotomaculum ruminis DNA as a template, dilute to 10 times according to a 10-fold equal gradient. -8 Gradient (concentrations are 121, 12.1, 1.21ng / μL, 121, 12.1, 1.21pg / μL, 121, 12.1, 1.21fg / μL). Different dilutions of DNA were used as PCR, LAMP and LAMP-LFD reaction templates.
[0055] 2. Use LAMP-LFD, LAMP and PCR methods to detect sulfate-reducing bacteria, and compare the sensitivity of the three methods.
[0056] 2.1 Amplify with the above-mentioned desulfovibrio (Desulfotomaculum ruminis) DNA through gradient dilution as the template of the LAMP reaction, according to the primers in the example 1 and the reaction system and temperature and time in the example 2, analyze the LAMP with the LFD method provided by the invention reaction product, see figure 1 .
[0057] 2.2 Amplify the above-mentioned...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com